1ITL
 
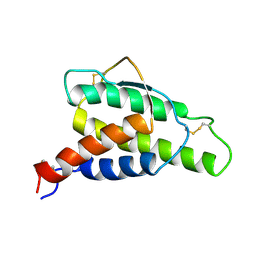 | | HUMAN INTERLEUKIN 4: THE SOLUTION STRUCTURE OF A FOUR-HELIX-BUNDLE PROTEIN | | Descriptor: | INTERLEUKIN-4 | | Authors: | Smith, L.J, Redfield, C, Boyd, J, Lawrence, G.M.P, Edwards, R.G, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1992-02-08 | | Release date: | 1993-04-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Human interleukin 4. The solution structure of a four-helix bundle protein.
J.Mol.Biol., 224, 1992
|
|
8RQP
 
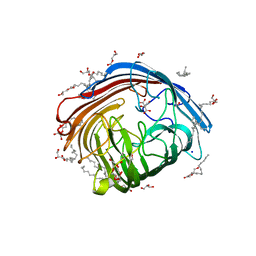 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), Alginate production protein AlgE, ... | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQR
 
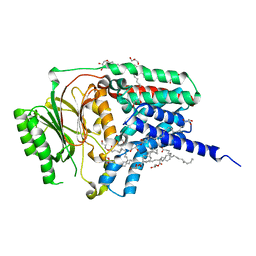 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (S-form), Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQQ
 
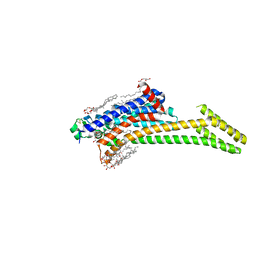 | | In meso structure of the adenosine A2a G protein-coupled receptor, A2aR, in 7.10 monoacylglycerol | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), ... | | Authors: | Smithers, L, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
7ACI
 
 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 9.8 monoacylglycerol | | Descriptor: | Apolipoprotein N-acyltransferase, GLYCEROL, [(2~{S})-2,3-bis(oxidanyl)propyl] heptadec-9-enoate | | Authors: | Smithers, L, van Dalsen, L, Boland, C, Caffrey, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
Cryst.Growth Des., 2020
|
|
7ACG
 
 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 9.8 monoacylglycerol | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-(2-METHOXYETHOXY)ETHANOL, Alginate biosynthesis protein AlgE, ... | | Authors: | Smithers, L, van Dalsen, L, Boland, C, Caffrey, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
Cryst.Growth Des., 2020
|
|
6G7G
 
 | | Structure of SPH (Self-Incompatibility Protein Homologue) proteins, a widespread family of small, highly stable, secreted proteins from plants | | Descriptor: | S-protein homolog 15 | | Authors: | Rajasekar, K.V, Coulthard, R.J, Ride, J.P, Ji, S, Winn, P.J, Wheeler, M.P, Hyde, E.I, Smith, L.J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-03-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of SPH (self-incompatibility protein homologue) proteins: a widespread family of small, highly stable, secreted proteins.
Biochem.J., 476, 2019
|
|
7ZGW
 
 | |
6HUE
 
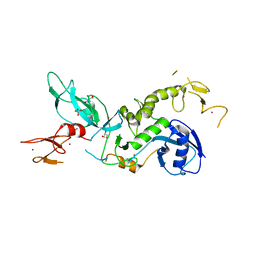 | | ParkinS65N | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | McWilliams, T.G, Barini, E, Pohjolan-Pirhonen, R, Brooks, S.P, Singh, F, Burel, S, Balk, K, Kumar, A, Montava-Garriga, L, Prescott, A.R, Hassoun, S.M, Mouton-Liger, F, Ball, G, Hills, R, Knebel, A, Ulusoy, A, Di Monte, D.A, Tamjar, J, Antico, O, Fears, K, Smith, L, Brambilla, R, Palin, E, Valori, M, Eerola-Rautio, J, Tienari, P, Corti, O, Dunnett, S.B, Ganley, I.G, Suomalainen, A, Muqit, M.M.K. | | Deposit date: | 2018-10-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Phosphorylation of Parkin at serine 65 is essential for its activation in vivo .
Open Biology, 8, 2018
|
|
7ZGV
 
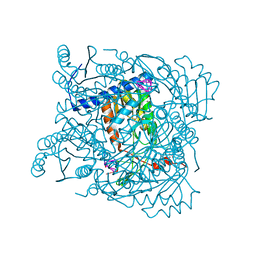 | | Serratia NucC bound to cA3 | | Descriptor: | ACETATE ION, CALCIUM ION, RNA (5'-R(P*AP*AP*A)-3'), ... | | Authors: | Garcia-Doval, C, Mayo-Munoz, D, Smith, L.M, Fineran, P.C. | | Deposit date: | 2022-04-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Type III CRISPR-Cas provides resistance against nucleus-forming jumbo phages via abortive infection.
Mol.Cell, 82, 2022
|
|
1ITM
 
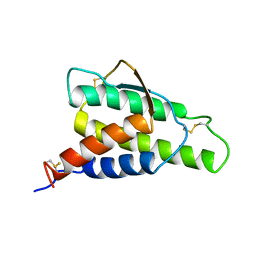 | | ANALYSIS OF THE SOLUTION STRUCTURE OF HUMAN INTERLEUKIN 4 DETERMINED BY HETERONUCLEAR THREE-DIMENSIONAL NUCLEAR MAGNETIC RESONANCE TECHNIQUES | | Descriptor: | INTERLEUKIN-4 | | Authors: | Redfield, C, Smith, L.J, Boyd, J, Lawrence, G.M.P, Edwards, R.G, Gershater, C.J, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1994-02-28 | | Release date: | 1994-05-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Analysis of the solution structure of human interleukin-4 determined by heteronuclear three-dimensional nuclear magnetic resonance techniques.
J.Mol.Biol., 238, 1994
|
|
4JMG
 
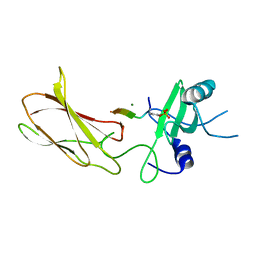 | |
4JMH
 
 | |
1E8L
 
 | | NMR solution structure of hen lysozyme | | Descriptor: | LYSOZYME | | Authors: | Schwalbe, H, Grimshaw, S.B, Spencer, A, Buck, M, Boyd, J, Dobson, C.M, Redfield, C, Smith, L.J. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | A refined solution structure of hen lysozyme determined using residual dipolar coupling data.
Protein Sci., 10, 2001
|
|
3PMW
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | DIMETHYL SULFOXIDE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PMV
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PMX
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1Z0H
 
 | | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Jayaraman, S, Eswarmoorthy, S, Ashraf, S.A, Smith, L.A, Swaminathan, S. | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
3O28
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-({[3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O2A
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O6I
 
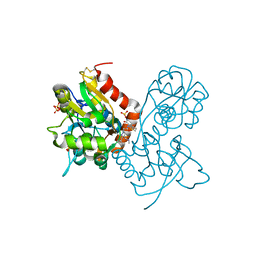 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-[({3-tert-butyl-4-[(methylamino)methyl]-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O29
 
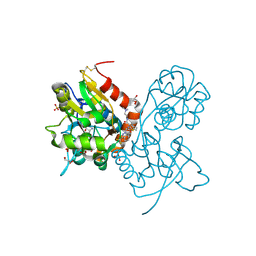 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O6H
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, 2-[({4-[(ethylamino)methyl]-3-(trifluoromethyl)-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O6G
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
