1DBI
 
 | | CRYSTAL STRUCTURE OF A THERMOSTABLE SERINE PROTEASE | | Descriptor: | AK.1 SERINE PROTEASE, CALCIUM ION, SODIUM ION | | Authors: | Smith, C.A, Toogood, H.S, Baker, H.M, Daniel, R.M, Baker, E.N. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calcium-mediated thermostability in the subtilisin superfamily: the crystal structure of Bacillus Ak.1 protease at 1.8 A resolution.
J.Mol.Biol., 294, 1999
|
|
8TH2
 
 | | Structure of the isoflavene-forming dirigent protein PsPTS2 | | Descriptor: | Dirigent protein | | Authors: | Smith, C.A, Meng, Q, Lewis, N.G, Davin, L.B. | | Deposit date: | 2023-07-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dirigent isoflavene-forming PsPTS2: 3D structure, stereochemical, and kinetic characterization comparison with pterocarpan-forming PsPTS1 homolog in pea.
J.Biol.Chem., 300, 2024
|
|
8V9H
 
 | | GES-5-NA-1-157 complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
8V9G
 
 | | GES-5-meropenem complex | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
8FAJ
 
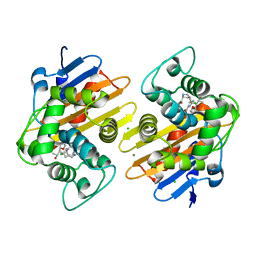 | | OXA-48-NA-1-157 inhibitor complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Toth, M, Vakulenko, S.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Exhibits Potent Activity against Klebsiella spp. Isolates Producing OXA-48-Type Carbapenemases.
Acs Infect Dis., 9, 2023
|
|
258D
 
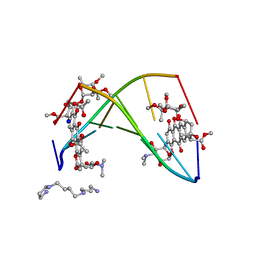 | | FACTORS AFFECTING SEQUENCE SELECTIVITY ON NOGALAMYCIN INTERCALATION: THE CRYSTAL STRUCTURE OF D(TGTACA)-NOGALAMYCIN | | Descriptor: | ACETATE ION, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3'), NOGALAMYCIN, ... | | Authors: | Smith, C.K, Brannigan, J.A, Moore, M.H. | | Deposit date: | 1996-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Factors affecting DNA sequence selectivity of nogalamycin intercalation: the crystal structure of d(TGTACA)2-nogalamycin2.
J.Mol.Biol., 263, 1996
|
|
7TVL
 
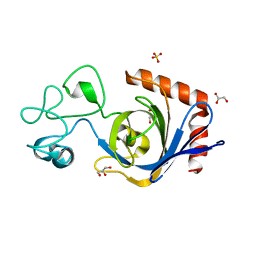 | | Viral AMG chitosanase V-Csn, apo structure | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVN
 
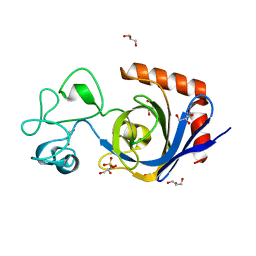 | | Viral AMG chitosanase V-Csn, D148N mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn D148N mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVO
 
 | | Viral AMG chitosanase V-Csn, E157Q mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn E157Q mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVP
 
 | | Viral AMG chitosanase V-Csn, E157Q mutant, chitotriose complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, GLYCEROL, Viral chitosanase V-Csn E157Q mutant chitotriose complex | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVM
 
 | | Viral AMG chitosanase V-Csn, apo structure, crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
6W5F
 
 | | Class D beta-lactamase BSU-2 delta mutant | | Descriptor: | 1,2-ETHANEDIOL, BSU-2delta mutant, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5O
 
 | | Class D beta-lactamase BAT-2 delta mutant | | Descriptor: | 1,2-ETHANEDIOL, BAT-2 Beta-lactamase delta mutant, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5E
 
 | | Class D beta-lactamase BSU-2 | | Descriptor: | 1,2-ETHANEDIOL, BSU-2 beta-lactamase, MALONATE ION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5G
 
 | | Class D beta-lactamase BAT-2 | | Descriptor: | 1,2-ETHANEDIOL, BAT-2 beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
5IY2
 
 | | Structure of apo OXA-143 carbapenemase | | Descriptor: | Beta-lactamase OXA-143, GLYCEROL | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2016-03-23 | | Release date: | 2017-08-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The role of conserved surface hydrophobic residues in the carbapenemase activity of the class D beta-lactamases.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1MND
 
 | |
7USZ
 
 | | Human DDAH-1, holo (Zn-bound) form | | Descriptor: | CHLORIDE ION, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, ZINC ION | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7UT0
 
 | | Human DDAH-1, apo form | | Descriptor: | 1,2-ETHANEDIOL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
1LCF
 
 | | CRYSTAL STRUCTURE OF COPPER-AND OXALATE-SUBSTITUTED HUMAN LACTOFERRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, COPPER (II) ION, ... | | Authors: | Smith, C.A, Anderson, B.F, Baker, H.M, Baker, E.N. | | Deposit date: | 1994-01-11 | | Release date: | 1994-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of copper- and oxalate-substituted human lactoferrin at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
8THA
 
 | | 1TEL, non-compressed, double-helical crystal form | | Descriptor: | Transcription factor ETV6,Activated CDC42 kinase 1 | | Authors: | Smith, C.P, Wilson, E.W, Pedroza Romo, M.J, Averett, J.C, Moody, J.D. | | Deposit date: | 2023-07-14 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 1TEL, non-compressed, double-helical crystal form
To Be Published
|
|
1MNE
 
 | |
182D
 
 | | DNA-NOGALAMYCIN INTERACTIONS: THE CRYSTAL STRUCTURE OF D(TGATCA) COMPLEXED WITH NOGALAMYCIN | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Smith, C.K, Davies, G.J, Dodson, E.J, Moore, M.H. | | Deposit date: | 1994-07-28 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA-nogalamycin interactions: the crystal structure of d(TGATCA) complexed with nogalamycin.
Biochemistry, 34, 1995
|
|
6OOC
 
 | | Structure of the pterocarpan synthase dirigent protein GePTS1 | | Descriptor: | Dirigent protein | | Authors: | Smith, C.A. | | Deposit date: | 2019-04-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pterocarpan synthase (PTS) structures suggest a common quinone methide-stabilizing function in dirigent proteins and proteins with dirigent-like domains.
J.Biol.Chem., 295, 2020
|
|
6PQC
 
 | | Structure of cefotaxime-CDD-1 beta-lactamase complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of CDD-1, the intrinsic class D beta-lactamase from the pathogenic Gram-positive bacterium Clostridioides difficile, and its complex with cefotaxime.
J.Struct.Biol., 208, 2019
|
|
