4NIK
 
 | |
5GVI
 
 | | Zebrafish USP30 in complex with Lys6-linked diubiquitin | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, ubiquitin | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30
Nat. Struct. Mol. Biol., 24, 2017
|
|
7E62
 
 | | Mouse TAB2 NZF in complex with Lys6-linked diubiquitin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGF-beta-activated kinase 1 and MAP3K7-binding protein 2, Ubiquitin, ... | | Authors: | Sato, Y, Li, Y, Okatsu, K, Fukai, S. | | Deposit date: | 2021-02-21 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for specific recognition of K6-linked polyubiquitin chains by the TAB2 NZF domain.
Biophys.J., 120, 2021
|
|
6JWI
 
 | | Yeast Npl4 in complex with Lys48-linked diubiquitin | | Descriptor: | BICINE, Nuclear protein localization protein 4, Ubiqutin, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-04-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
6KC6
 
 | | HOIP-HOIPIN8 complex | | Descriptor: | 2-[3-[2,6-bis(fluoranyl)-4-(1~{H}-pyrazol-4-yl)phenyl]-3-oxidanylidene-propyl]-4-(1-methylpyrazol-4-yl)benzoic acid, CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-06-27 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses.
Commun Biol, 3, 2020
|
|
6KC5
 
 | | HOIP-HOIPIN1 complex | | Descriptor: | 2-[3-(2-methoxyphenyl)-3-oxidanylidene-propyl]benzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RNF31, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-06-27 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses.
Commun Biol, 3, 2020
|
|
3A9E
 
 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | Authors: | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-10-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
6JWJ
 
 | | Npl4 in complex with Ufd1 | | Descriptor: | GLYCEROL, Nuclear protein localization protein 4, Peptide from Ubiquitin fusion degradation protein 1, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-04-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
6JWH
 
 | | Yeast Npl4 zinc finger, MPN and CTD domains | | Descriptor: | GLYCEROL, Nuclear protein localization protein 4, ZINC ION | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-04-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.72000253 Å) | | Cite: | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
6M35
 
 | | Crystal structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sato, Y, Yabuki, T, Arakawa, T, Yamada, C, Fushinobu, S, Wakagi, T. | | Deposit date: | 2020-03-02 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
3A2M
 
 | | CRYSTAL STRUCTURE OF DBJA (WILD TYPE Type I) | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Sato, Y, Senda, T. | | Deposit date: | 2009-05-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6IES
 
 | | Onion lachrymatory factor synthase (LFS) containing (E)-2-propen 1-ol (crotyl alcohol) | | Descriptor: | (2E)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Sato, Y, Arakawa, T, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase.
Acs Catalysis, 10, 2020
|
|
5YJE
 
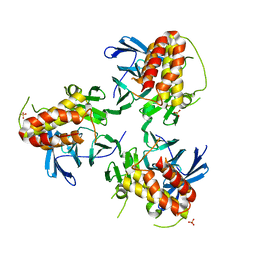 | | Crystal structure of HIRA(644-1017) | | Descriptor: | Protein HIRA, SULFATE ION | | Authors: | Sato, Y, Senda, M, Senda, T. | | Deposit date: | 2017-10-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit
Nat Commun, 9, 2018
|
|
2ZNV
 
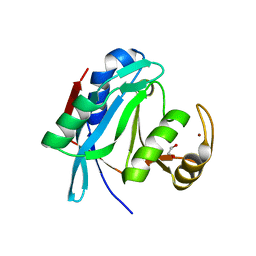 | | Crystal structure of human AMSH-LP DUB domain in complex with Lys63-linked ubiquitin dimer | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, Ubiquitin, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
2ZNR
 
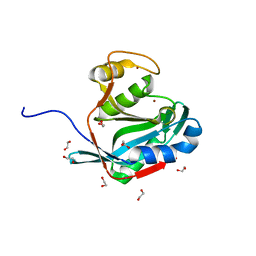 | | Crystal structure of the DUB domain of human AMSH-LP | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, PRASEODYMIUM ION, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
2ZXZ
 
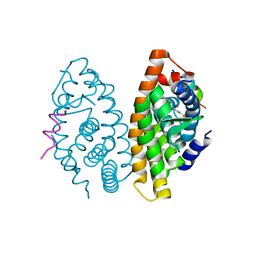 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-inden-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
3A2L
 
 | | Crystal structure of DBJA (mutant dbja delta) | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Sato, Y, Senda, T. | | Deposit date: | 2009-05-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3AFI
 
 | | Crystal structure of DBJA (HIS-DBJA) | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Sato, Y, Senda, T. | | Deposit date: | 2010-03-02 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3A2N
 
 | | Crystal structure of DBJA (Wild Type Type II P21) | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Sato, Y, Senda, T. | | Deposit date: | 2009-05-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3AJB
 
 | | Crystal Structure of human Pex3p in complex with N-terminal Pex19p peptide | | Descriptor: | Peroxisomal biogenesis factor 19, Peroxisomal biogenesis factor 3 | | Authors: | Sato, Y, Shibata, H, Nakatsu, T, Kato, H. | | Deposit date: | 2010-05-27 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p
Embo J., 29, 2010
|
|
6M3X
 
 | | Cryo-EM structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, Sulfur oxygenase/reductase | | Authors: | Sato, Y, Adachi, N, Moriya, T, Arakawa, T, Kawasaki, M, Yamada, C, Senda, T, Fushinobu, S. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
3WXG
 
 | |
3WXE
 
 | |
3A9J
 
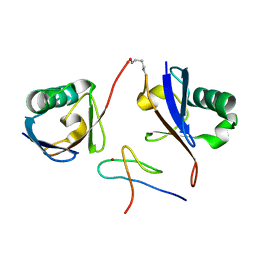 | | Crystal structure of the mouse TAB2-NZF in complex with Lys63-linked di-ubiquitin | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2, Ubiquitin, ZINC ION | | Authors: | Sato, Y, Yoshikawa, A, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3
Embo J., 28, 2009
|
|
3WWQ
 
 | |
