4H7B
 
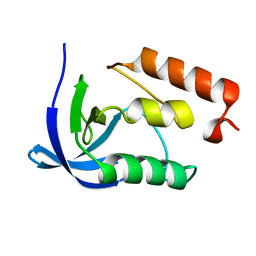 | |
4K8I
 
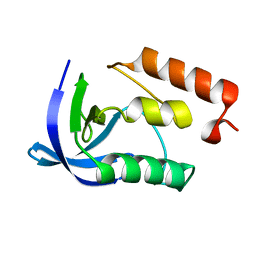 | | Crystal Structure of Staphylococcal Nuclease mutant I92V/V99L | | Descriptor: | Thermonuclease | | Authors: | Sanders, J.M, Latimer, E.C, Roeser, J.R, Janowska, K, Sakon, J, Stites, W.E. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic core mutants of Staphylococcal nuclease
To be Published
|
|
4K8J
 
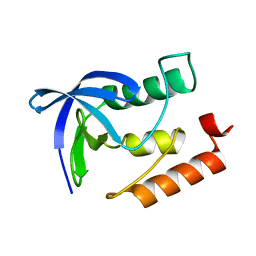 | | Crystal Structure of Staphylococcal nuclease mutant V23L/V66I | | Descriptor: | Thermonuclease | | Authors: | Sanders, J.M, Gill, E, Roeser, J.R, Janowska, K, Sakon, J, Stites, W.E. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core mutants of Staphylococcal nuclease
To be Published
|
|
4XUC
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd18 (1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one) | | Descriptor: | 1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
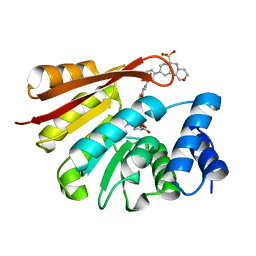
4XUD
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd32 ([1-(biphenyl-3-yl)-5-hydroxy-4-oxo-1,4-dihydropyridin-3-yl]boronic acid) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4XUE
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd27b | | Descriptor: | 2-(biphenyl-3-yl)-5-hydroxy-3-methylpyrimidin-4(3H)-one, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4HHE
 
 | | Quinolinate synthase from Pyrococcus furiosus | | Descriptor: | CHLORIDE ION, Quinolinate synthase A | | Authors: | Soriano, E.V, Zhang, Y, Settembre, E.C, Colabroy, K, Sanders, J.M, Dorrestein, P.C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-10-09 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Active-site models for complexes of quinolinate synthase with substrates and intermediates.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6NM4
 
 | | Crystal structure of SAM-bound PRDM9 in complex with MRK-740 inhibitor | | Descriptor: | 4-[3-(3,5-dimethoxyphenyl)-1,2,4-oxadiazol-5-yl]-1-methyl-9-(2-methylpyridin-4-yl)-1,4,9-triazaspiro[5.5]undecane, Histone-lysine N-methyltransferase PRDM9, S-ADENOSYLMETHIONINE, ... | | Authors: | Ivanochko, D, Halabelian, L, Fischer, C, Sanders, J.M, Kattar, S.D, Brown, P.J, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of a chemical probe for PRDM9.
Nat Commun, 10, 2019
|
|
6VLM
 
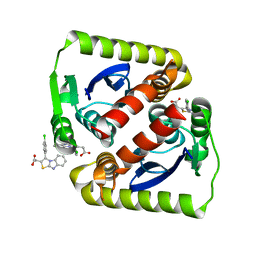 | |
5U6I
 
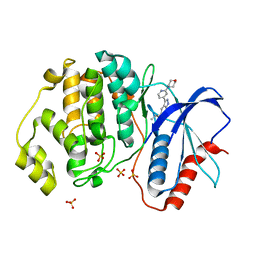 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
1T1R
 
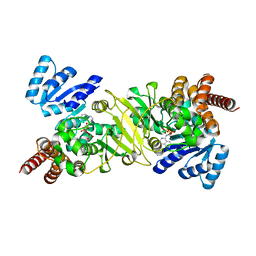 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, SULFATE ION, [(ISOQUINOLIN-1-YLAMINO)-PHOSPHONO-METHYL]-PHOSPHONIC ACID | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
1TGY
 
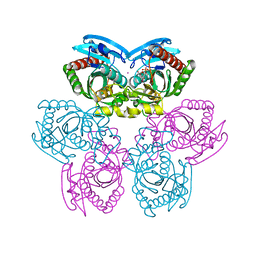 | | Structure of E. coli Uridine Phosphorylase complexed with uracil and ribose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, POTASSIUM ION, URACIL, ... | | Authors: | Bu, W, Settembre, E.C, Sanders, J.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of E. coli Uridine Phosphorylase
To be Published, 2004
|
|
1T1S
 
 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
1TGV
 
 | | Structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate | | Descriptor: | 5-FLUOROURIDINE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Bu, W, Settembre, E.C, Sanders, J.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of E. coli Uridine Phosphorylase
To be Published, 2004
|
|
