5CZK
 
 | | Structure of E. coli beta-glucuronidase bound with a novel, potent inhibitor 1-((6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea | | Descriptor: | 1-[(6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Roberts, A.R, Wallace, B.R, Redinbo, M.R. | | Deposit date: | 2015-07-31 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity.
Chem.Biol., 22, 2015
|
|
1RLM
 
 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLT
 
 | | Transition State Analogue of ybiV from E. coli K12 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLO
 
 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
4HTG
 
 | | Porphobilinogen Deaminase from Arabidopsis Thaliana | | Descriptor: | 3-[(5Z)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methylidene}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, ACETATE ION, Porphobilinogen deaminase, ... | | Authors: | Roberts, A, Gill, R, Hussey, R.J, Erskine, P.T, Cooper, J.B, Wood, S.P, Chrystal, E.J.T, Shoolingin-Jordan, P.M. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHZ
 
 | | Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide | | Descriptor: | 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide, Beta-glucuronidase | | Authors: | Roberts, A.B, Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Molecular Insights into Microbial beta-Glucuronidase Inhibition to Abrogate CPT-11 Toxicity.
Mol.Pharmacol., 84, 2013
|
|
2G7H
 
 | |
4UX7
 
 | | Structure of a Clostridium difficile sortase | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PUTATIVE PEPTIDASE C60B, ... | | Authors: | Chambers, C.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2014-08-19 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Function of a Clostridium Difficile Sortase Enzyme.
Sci.Rep., 5, 2015
|
|
7NB8
 
 | | Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-5, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
7NBA
 
 | | Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin motor domain-containing protein,Kinesin motor domain-containing protein, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
8BBF
 
 | | Structure of the IFT-A complex; IFT-A1 module | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, WD repeat-containing protein 19 | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
8BBE
 
 | | Structure of the IFT-A complex; IFT-A2 module | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 43 homolog, SNAP-tag,Tetratricopeptide repeat protein 21B, ... | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
8BBG
 
 | | Structure of the IFT-A complex; anterograde IFT-A train model | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
8RGG
 
 | | Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Dynein light chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Mladenov, M, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGI
 
 | | Structure of DYNLT1:DYNLT2B (TCTEX1:TCTEX1D2) heterodimer. | | Descriptor: | Dynein light chain Tctex-type 1, Dynein light chain Tctex-type protein 2B | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGH
 
 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
5NJL
 
 | | Cwp2 from Clostridium difficile | | Descriptor: | Cell surface protein (Putative S-layer protein), SULFATE ION | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-03-29 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cwp2 from Clostridium difficile exhibits an extended three domain fold and cell adhesion in vitro.
FEBS J., 284, 2017
|
|
5M54
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-J, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5M5C
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5M50
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2WOT
 
 | | ALK5 IN COMPLEX WITH 4-((5,6-dimethyl-2-(2-pyridyl)-3-pyridyl)oxy)-N-(3,4,5-trimethoxyphenyl)pyridin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5,6-DIMETHYL-2,2'-BIPYRIDIN-3-YL)OXY]-N-(3,4,5-TRIMETHOXYPHENYL)PYRIDIN-2-AMINE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Norman, R.A, Debreczeni, J.E, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
2WOU
 
 | | ALK5 IN COMPLEX WITH 4-((4-((2,6-dimethyl-3-pyridyl)oxy)-2-pyridyl) amino)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(2,6-DIMETHYLPYRIDIN-3-YL)OXY]PYRIDIN-2-YL}AMINO)BENZENESULFONAMIDE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Debreczeni, J.E, Norman, R.A, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
5OQ3
 
 | | High resolution structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cwp19, ... | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
5OQ2
 
 | | Se-SAD structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, Cwp19, PHOSPHATE ION | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
6RLA
 
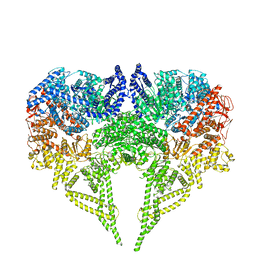 | | Structure of the dynein-2 complex; motor domains | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Toropova, K, Zalyte, R, Mukhopadhyay, A.G, Mladenov, M, Carter, A.P, Roberts, A.J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Nat.Struct.Mol.Biol., 26, 2019
|
|
