6QPM
 
 | | Adenovirus serotype 10 Fiber-Knob | | Descriptor: | Fiber protein | | Authors: | Baker, A.T, Rizkallah, P.J. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Development of a low-seroprevalence, alpha v beta 6 integrin-selective virotherapy based on human adenovirus type 10.
Mol Ther Oncolytics, 25, 2022
|
|
8BAD
 
 | | Tpp80Aa1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Binary toxin A-like protein, CALCIUM ION, ... | | Authors: | Best, H.L, Williamson, L.J, Rizkallah, P.J, Berry, C, Lloyd-Evans, E. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure of Bacillus thuringiensis Tpp80Aa1 and Its Interaction with Galactose-Containing Glycolipids.
Toxins, 14, 2022
|
|
6QPN
 
 | | Adenovirus species D serotype 49 Fiber-Knob | | Descriptor: | Fiber, SULFATE ION | | Authors: | Baker, A.T, Rizkallah, P.J. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The Fiber Knob Protein of Human Adenovirus Type 49 Mediates
Highly Efficient and Promiscuous Infection of Cancer Cell Lines
Using a Novel Cell Entry Mechanism
Journal of Virology, 95, 2021
|
|
6QZA
 
 | | HLA-DR1 with GMF Influenza PB1 Peptide | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Greenshields-Watson, A, Rizkallah, P.J. | | Deposit date: | 2019-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | CD4+T Cells Recognize Conserved Influenza A Epitopes through Shared Patterns of V-Gene Usage and Complementary Biochemical Features.
Cell Rep, 32, 2020
|
|
6QPO
 
 | | Adenovirus species D serotype 49 Fiber-Knob KO1 mutant | | Descriptor: | Fiber, SULFATE ION | | Authors: | Baker, A.T, Rizkallah, P.J. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Fiber Knob Protein of Human Adenovirus Type 49 Mediates Highly Efficient and Promiscuous Infection of Cancer Cell Lines Using a Novel Cell Entry Mechanism.
J.Virol., 95, 2021
|
|
6QZC
 
 | | HLA-DR1 with the QAR Peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Greenshields-Watson, A, Rizkallah, P.J. | | Deposit date: | 2019-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | CD4+T Cells Recognize Conserved Influenza A Epitopes through Shared Patterns of V-Gene Usage and Complementary Biochemical Features.
Cell Rep, 32, 2020
|
|
4PHN
 
 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids | | Descriptor: | CALCIUM ION, Calpain small subunit 1 | | Authors: | Allemann, R.K, Rizkallah, P.J, Adams, S.E, Miller, D.J, Hallett, M.B. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
4PHJ
 
 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids: Human unliganded protein | | Descriptor: | CALCIUM ION, Calpain small subunit 1 | | Authors: | Adams, S.E, Rizkallah, P.J, Allemann, R.K, Miller, D.J, Hallett, M.B, Robinson, E. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
1S44
 
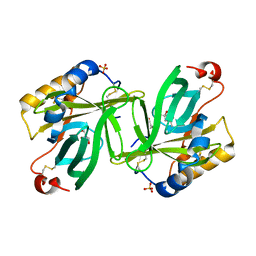 | | The structure and refinement of apocrustacyanin C2 to 1.6A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Crustacyanin A1 subunit, GLYCEROL, ... | | Authors: | Habash, J, Helliwell, J.R, Raftery, J, Cianci, M, Rizkallah, P.J, Chayen, N.E, Nneji, G.A, Zagalsky, P.F. | | Deposit date: | 2004-01-15 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure and refinement of apocrustacyanin C2 to 1.3 A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4J88
 
 | | Dark-state structure of sfGFP containing the unnatural amino acid p-azido-phenylalanine at residue 66 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Reddington, S.C, Jones, D.D, Rizkallah, P.J, Tippmann, E.M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Different Photochemical Events of a Genetically Encoded Phenyl Azide Define and Modulate GFP Fluorescence.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4J8A
 
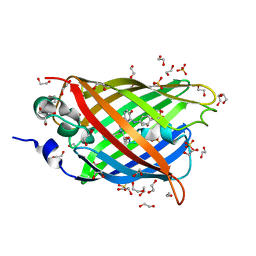 | | Irradiated-state structure of sfGFP containing the unnatural amino acid p-azido-phenylalanine at residue 145 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Reddington, S.C, Jones, D.D, Rizkallah, P.J, Tippmann, E.M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Different Photochemical Events of a Genetically Encoded Phenyl Azide Define and Modulate GFP Fluorescence.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5NHN
 
 | |
4J89
 
 | | Different photochemical events of a genetically encoded aryl azide define and modulate GFP fluorescence | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Reddington, S.C, Jones, D.D, Rizkallah, P.J, Tippmann, E.M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different Photochemical Events of a Genetically Encoded Phenyl Azide Define and Modulate GFP Fluorescence.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6HG6
 
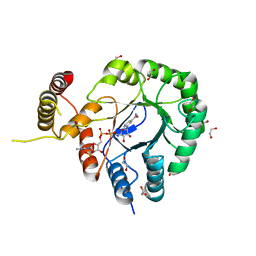 | | Clostridium beijerinckii aldo-keto reductase Cbei_3974 with NADPH | | Descriptor: | 1,2-ETHANEDIOL, L-glyceraldehyde 3-phosphate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scott, A.F, Cresser-Brown, J, Rizkallah, P.J, Jin, Y, Allemann, R.K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Biophysical Analysis of Furfural-Detoxifying Aldehyde Reductase from Clostridium beijerinckii.
Appl.Environ.Microbiol., 85, 2019
|
|
4KAG
 
 | | Crystal structure analysis of a single amino acid deletion mutation in EGFP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Arpino, J.A.J, Rizkallah, P.J. | | Deposit date: | 2013-04-22 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and dynamic changes associated with beneficial engineered single-amino-acid deletion mutations in enhanced green fluorescent protein.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KA9
 
 | |
4ZIO
 
 | | Irradiated state of mCherry143azF | | Descriptor: | SULFATE ION, mCherry | | Authors: | Reddington, S.C, Driezis, S, Hartley, A.M, Watson, P.D, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-04-28 | | Release date: | 2015-09-16 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genetically encoded phenyl azide photochemistry drives positive and negative functional modulation of a red fluorescent protein
Rsc Adv, 5, 2015
|
|
4ZIN
 
 | | Genetically encoded Phenyl Azide Photochemistry Drive Positive and Negative Functional Modulation of a Red Fluorescent Protein | | Descriptor: | 1,2-ETHANEDIOL, MCherry fluorescent protein, SULFATE ION | | Authors: | Reddington, S.C, Driezis, S, Hartley, A.M, Watson, P.D, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-04-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Genetically encoded phenyl azide photochemistry drives positive and negative functional modulation of a red fluorescent protein
Rsc Adv, 5, 2015
|
|
1KI2
 
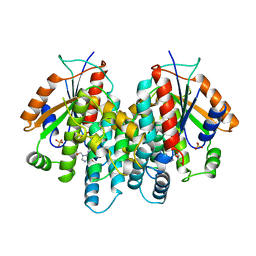 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH GANCICLOVIR | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KIM
 
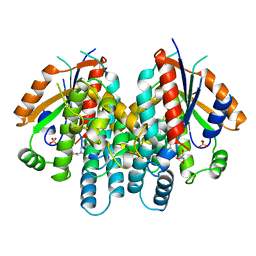 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH DEOXYTHYMIDINE | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
5BS0
 
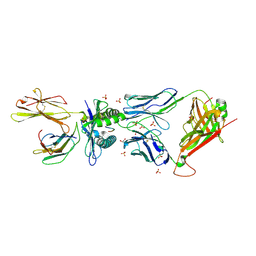 | | MAGE-A3 Reactive TCR in complex with Titin Epitope in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, Lomax, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
5BRZ
 
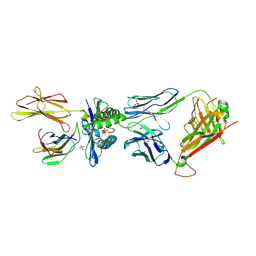 | | MAGE-A3 reactive TCR in complex with MAGE-A3 in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-PRO-ILE-GLY-HIS-LEU-TYR, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donnellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, LomaX, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
4KEX
 
 | |
5BTT
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | GLYCEROL, Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5BT0
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-02 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
