5ABP
 
 | |
4KRD
 
 | | Crystal Structure of Pho85-Pcl10 Complex | | Descriptor: | Cyclin-dependent protein kinase PHO85, PHO85 cyclin-10 | | Authors: | Quiocho, F.A, Zheng, F. | | Deposit date: | 2013-05-16 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | New Structural Insights into Phosphorylation-free Mechanism for Full Cyclin-dependent Kinase (CDK)-Cyclin Activity and Substrate Recognition.
J.Biol.Chem., 288, 2013
|
|
4KRC
 
 | | Crystal Structure of Pho85-Pcl10-ATP-gamma-S Complex | | Descriptor: | Cyclin-dependent protein kinase PHO85, MAGNESIUM ION, PHO85 cyclin-10, ... | | Authors: | Quiocho, F.A, Zheng, F. | | Deposit date: | 2013-05-16 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | New Structural Insights into Phosphorylation-free Mechanism for Full Cyclin-dependent Kinase (CDK)-Cyclin Activity and Substrate Recognition.
J.Biol.Chem., 288, 2013
|
|
2AQL
 
 | | Crystal Structure of the MRG15 MRG domain | | Descriptor: | Mortality factor 4-like protein 1 | | Authors: | Quiocho, F.A, Bowman, B.R. | | Deposit date: | 2005-08-18 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain.
Structure, 14, 2006
|
|
8ABP
 
 | |
7ABP
 
 | |
9ABP
 
 | |
6ABP
 
 | |
4MBP
 
 | | MALTODEXTRIN BINDING PROTEIN WITH BOUND MALTETROSE | | Descriptor: | MALTODEXTRIN BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-08-17 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
2GBP
 
 | |
2VP3
 
 | |
3MBP
 
 | | MALTODEXTRIN-BINDING PROTEIN WITH BOUND MALTOTRIOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
1BAP
 
 | |
1APB
 
 | |
2LIV
 
 | |
1CDM
 
 | |
1CLL
 
 | |
2LBP
 
 | |
4W9N
 
 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase complexed with triclosan | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, IMIDAZOLE, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
4W82
 
 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase | | Descriptor: | CHLORIDE ION, Fatty acid synthase, MAGNESIUM ION, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
2PFD
 
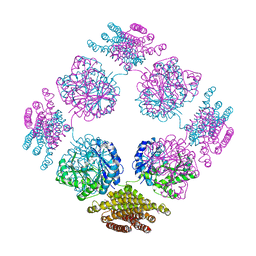 | | Anisotropically refined structure of FTCD | | Descriptor: | Formimidoyltransferase-cyclodeaminase | | Authors: | Poon, B.K, Chen, X, Lu, M, Quiocho, F.A, Wang, Q, Ma, J. | | Deposit date: | 2007-04-04 | | Release date: | 2007-04-24 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Anisotropically refined structure of FTCD
To be Published
|
|
1ABE
 
 | |
1ANF
 
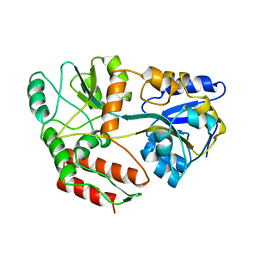 | | MALTODEXTRIN BINDING PROTEIN WITH BOUND MALTOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
2ADA
 
 | |
1DPE
 
 | | DIPEPTIDE-BINDING PROTEIN | | Descriptor: | CADMIUM ION, DIPEPTIDE-BINDING PROTEIN | | Authors: | Nickitenko, A.V, Trakhanov, S, Quiocho, F.A. | | Deposit date: | 1995-07-25 | | Release date: | 1996-08-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2 A resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor.
Biochemistry, 34, 1995
|
|
