5FGM
 
 | | Streptomyces coelicolor SigR region 4 | | Descriptor: | ECF RNA polymerase sigma factor SigR | | Authors: | Park, S.Y. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In Streptomyces coelicolor SigR, methionine at the -35 element interacting region 4 confers the -31'-adenine base selectivity
Biochem.Biophys.Res.Commun., 470, 2016
|
|
5X14
 
 | | Crystal structure of Bacillus subtilis PadR in complex with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X13
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
2KSJ
 
 | | Structure and Dynamics of the Membrane-bound form of Pf1 Coat Protein: Implications for Structural Rearrangement During Virus Assembly | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Marassi, F, Black, D, Opella, S.J. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly.
Biophys.J., 99, 2010
|
|
8KCA
 
 | |
8EY0
 
 | | Structure of an orthogonal PYR1*:HAB1* chemical-induced dimerization module in complex with mandipropamid | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, GLYCEROL, ... | | Authors: | Park, S.-Y, Volkman, B.F, Cutler, S.R, Peterson, F.C. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity.
Nat.Chem.Biol., 20, 2024
|
|
6PWW
 
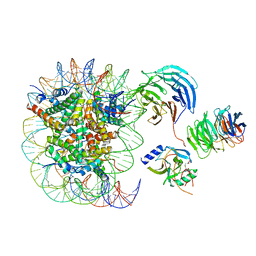 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWX
 
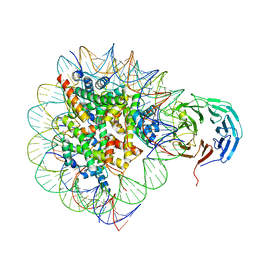 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
2KNH
 
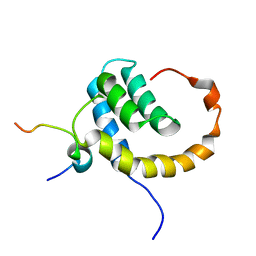 | | The Solution structure of the eTAFH domain of AML1-ETO complexed with HEB peptide | | Descriptor: | Protein CBFA2T1, Transcription factor 12 | | Authors: | Park, S, Cierpicki, T, Tonelli, M, Bushweller, J.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity.
Blood, 113, 2009
|
|
6W5N
 
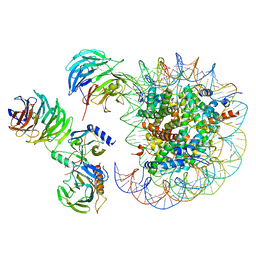 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5I
 
 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5M
 
 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
2KYU
 
 | | The solution structure of the PHD3 finger of MLL | | Descriptor: | Histone-lysine N-methyltransferase MLL, ZINC ION | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression.
Biochemistry, 49, 2010
|
|
5JE8
 
 | |
2KYX
 
 | | Solution structure of the RRM domain of CYP33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression .
Biochemistry, 49, 2010
|
|
5GV5
 
 | | Crystal structure of Candida antarctica Lipase B with active Ser105 modified with a phosphonate inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, ... | | Authors: | Park, S.Y, Lee, H. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural and Experimental Evidence for the Enantiomeric Recognition toward a Bulky sec-Alcohol by Candida antarctica Lipase B
Acs Catalysis, 6, 2016
|
|
5CSS
 
 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
5CSR
 
 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilium | | Descriptor: | CHLORIDE ION, GLYCEROL, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Park, H.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
8SQG
 
 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
4RDH
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Park, S.Y, Kim, S, Lee, H. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity.
J.Mol.Biol., 427, 2015
|
|
8SQF
 
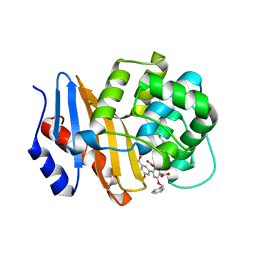 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
4RDI
 
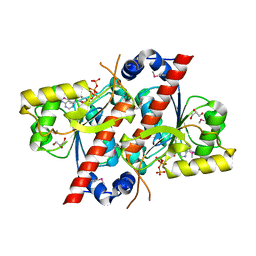 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, tRNA threonylcarbamoyladenosine dehydratase | | Authors: | Park, S.Y, Kim, S, Lee, H. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity.
J.Mol.Biol., 427, 2015
|
|
4N7V
 
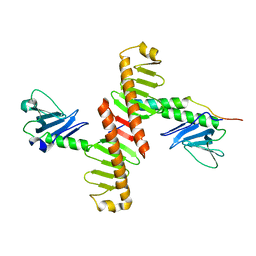 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep152 N-terminal fragment | | Descriptor: | Centrosomal protein of 152 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4N7Z
 
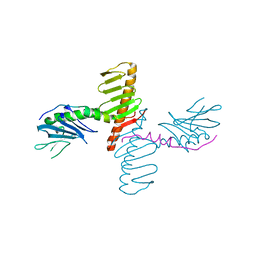 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep192 N-terminal fragment | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
