1XNY
 
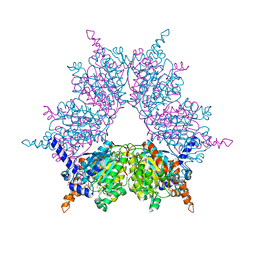 | | Biotin and propionyl-CoA bound to Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB) | | Descriptor: | BIOTIN, propionyl Coenzyme A, propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
1XNW
 
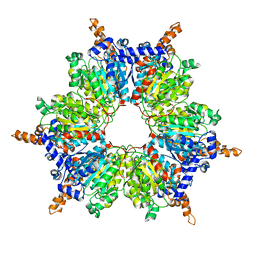 | | Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB), apo form #2, mutant D422I | | Descriptor: | propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
1XO6
 
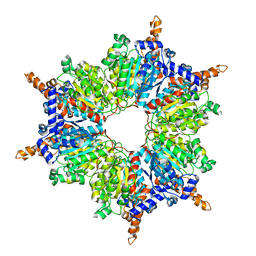 | | Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB), apo form #3 | | Descriptor: | propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
2VMB
 
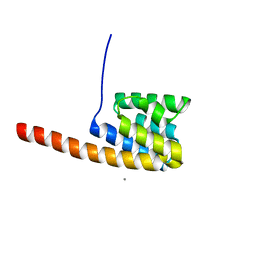 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
2VMA
 
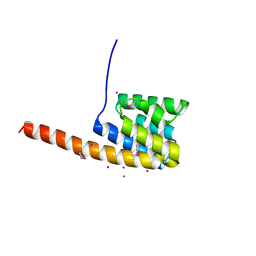 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F, IODIDE ION | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
7QW3
 
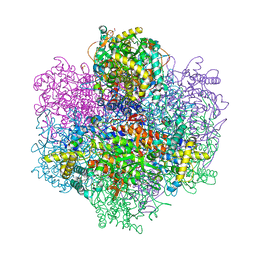 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with Br ion. | | Descriptor: | BROMIDE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QWI
 
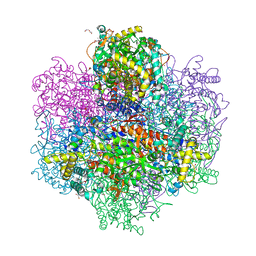 | | Vanadate complex of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QVW
 
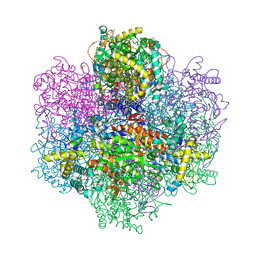 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QYY
 
 | | Vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with chloride | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-29 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
1XAR
 
 | | Crystal Structure of a fragment of DC-SIGNR (containing the carbohydrate recognition domain and two repeats of the neck). | | Descriptor: | CD209 antigen-like protein 1, SODIUM ION | | Authors: | Feinberg, H, Guo, Y, Mitchell, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2004-08-26 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Extended Neck Regions Stabilize Tetramers of the Receptors DC-SIGN and DC-SIGNR
J.Biol.Chem., 280, 2005
|
|
1XNV
 
 | | Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB), apo form #1 | | Descriptor: | propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SL4
 
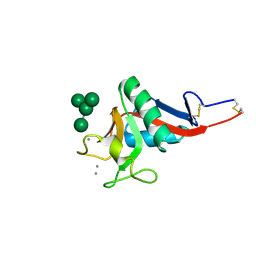 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SL6
 
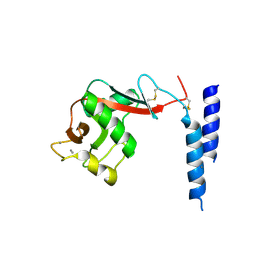 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TS2
 
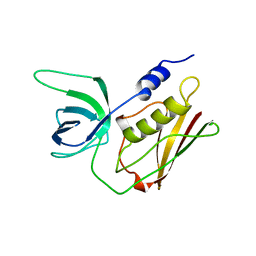 | | T128A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
1TS3
 
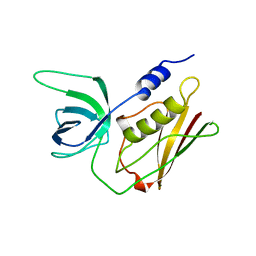 | | H135A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
1K9J
 
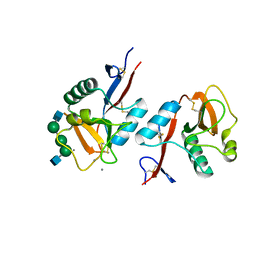 | | Complex of DC-SIGNR and GlcNAc2Man3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, mDC-SIGN2 TYPE I ISOFORM | | Authors: | Feinberg, H, Mitchell, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2001-10-29 | | Release date: | 2001-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR.
Science, 294, 2001
|
|
1K9I
 
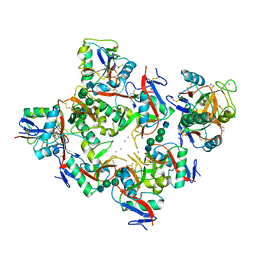 | | Complex of DC-SIGN and GlcNAc2Man3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, mDC-SIGN1B type I isoform | | Authors: | Feinberg, H, Mitchell, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2001-10-29 | | Release date: | 2001-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR.
Science, 294, 2001
|
|
1RDP
 
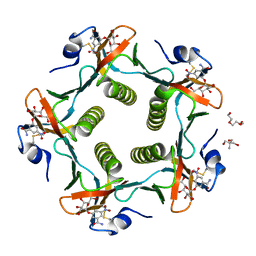 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RF2
 
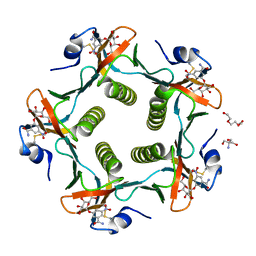 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV4 | | Descriptor: | 1,3-BIS-([3-[3-[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBUTENYL]-AMINO-PROPOXY-ETHOXY-ETHOXY]-PROPYL-]AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-07 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RD9
 
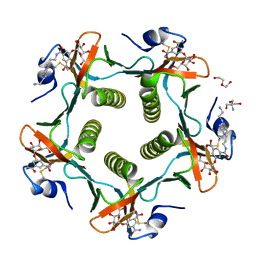 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV2 | | Descriptor: | 1,3-BIS-([3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYL-AMINO]-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RCV
 
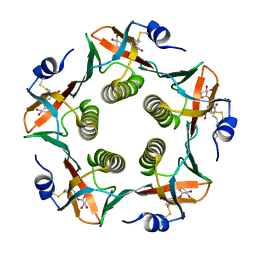 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV1 | | Descriptor: | [3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO] -2-(3-{4-[3-(3-NITRO-5-[GALACTOPYRANOSYLOXY]-BENZOYLAMINO)-PROPYL]-PIPERAZIN-1-YL} -PROPYL-AMINO)-3,4-DIOXO-CYCLOBUTENE, cholera toxin B protein (CTB) | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-04 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
2MW3
 
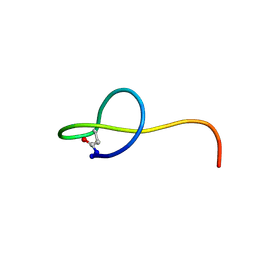 | | Solution NMR structure of the lasso peptide streptomonomicin | | Descriptor: | Lasso peptide | | Authors: | Tietz, J.I, Zhu, L, Mitchell, D.A, Metelev, M, Melby, J.O, Blair, P.M, Livnat, I, Severinov, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-01-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso Peptide from an understudied halophilic actinomycete.
Chem.Biol., 22, 2015
|
|
1LVZ
 
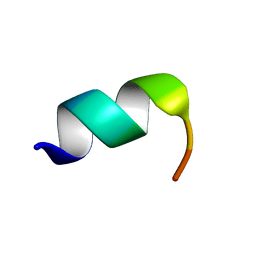 | | METARHODOPSIN II BOUND STRUCTURE OF C-TERMINAL PEPTIDE OF ALPHA-SUBUNIT OF TRANSDUCIN | | Descriptor: | Guanine nucleotide-binding protein G(T), alpha-1 subunit | | Authors: | Koenig, B.W, Kontaxis, G, Mitchell, D.C, Louis, J.M, Litman, B.J, Bax, A. | | Deposit date: | 2002-05-30 | | Release date: | 2002-09-11 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of a G protein fragment in the receptor bound state from residual dipolar couplings.
J.Mol.Biol., 322, 2002
|
|
1HLM
 
 | |
