2DP5
 
 | | Structure of streptococcus pyogenes bacteriophage-associated hyaluronate lyase Hylp2 | | Descriptor: | Hyaluronidase | | Authors: | Mishra, P, Bhakuni, V, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-05-06 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of streptococcus pyogenes bacteriophage-associated hyaluronate lyase Hylp2
To be Published
|
|
2YVV
 
 | | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites | | Descriptor: | Hyaluronidase, phage associated, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mishra, P, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Bhakuni, V, Singh, T.P. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites
To be Published
|
|
2YX2
 
 | | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Mishra, P, Prem Kumar, R, Bhakuni, V, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution
To be Published
|
|
3EKA
 
 | | Crystal structure of the complex of hyaluranidase trimer with ascorbic acid at 3.1 A resolution reveals the locations of three binding sites | | Descriptor: | ASCORBIC ACID, Hyaluronidase, phage associated | | Authors: | Mishra, P, Ethayathulla, A.S, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Bhakuni, V, Singh, T.P. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose.
Febs J., 276, 2009
|
|
1JXJ
 
 | | Role of mobile loop in the mechanism of human salivary amylase | | Descriptor: | Alpha-amylase, salivary, CALCIUM ION, ... | | Authors: | Ramasubbu, N, Ragunath, C, Wang, Z, Mishra, P.J, Thomas, L.M. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human salivary alpha-amylase Trp58 situated at subsite -2 is critical for enzyme activity.
Eur.J.Biochem., 271, 2004
|
|
1Z32
 
 | | Structure-function relationships in human salivary alpha-amylase: Role of aromatic residues | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, CALCIUM ION, ... | | Authors: | Ramasubbu, N, Ragunath, C, Sundar, K, Mishra, P.J, Gyemant, G, Kandra, L. | | Deposit date: | 2005-03-10 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function relationships in human salivary alpha-amylase: Role of aromatic residues
To be Published
|
|
2YW0
 
 | | Crystal structure of hyluranidase trimer at 2.6 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Prem Kumar, R, Mishra, P, Singh, N, Perbandt, M, Kaur, P, Sharma, S, Betzel, C, Bhakuni, V, Singh, T.P. | | Deposit date: | 2007-04-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose
Febs J., 276, 2009
|
|
2Q2L
 
 | | Crystal Structure of Superoxide Dismutase from P. atrosanguina | | Descriptor: | IODIDE ION, Superoxide Dismutase, ZINC ION | | Authors: | Manickam, Y, Gill, J, Mishra, P.C, Sharma, A. | | Deposit date: | 2007-05-29 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | SAD phasing of a structure based on cocrystallized iodides using an in-house Cu Kalpha X-ray source: effects of data redundancy and completeness on structure solution
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1MFU
 
 | | Probing the role of a mobile loop in human salivary amylase: Structural studies on the loop-deleted mutant | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Ramasubbu, N, Ragunath, C, Mishra, P.J. | | Deposit date: | 2002-08-13 | | Release date: | 2002-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of a mobile loop in substrate binding and enzyme activity of
human salivary amylase.
J.Mol.Biol., 325, 2003
|
|
1NM9
 
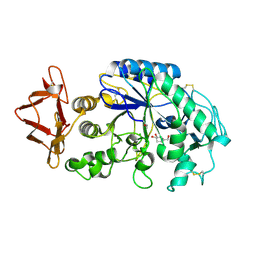 | | Crystal structure of recombinant human salivary amylase mutant W58A | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, Alpha-amylase, ... | | Authors: | Ramasubbu, N, Ragunath, C, Mishra, P.J, Thomas, L.M. | | Deposit date: | 2003-01-09 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human salivary alpha-amylase Trp58 situated at subsite -2 is critical for enzyme activity.
Eur.J.Biochem., 271, 2004
|
|
1MFV
 
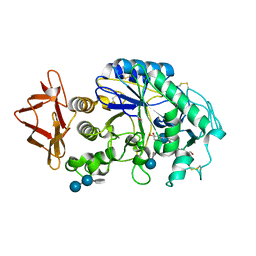 | | Probing the role of a mobile loop in human slaivary amylase: Structural studies on the loop-deleted enzyme | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Ramasubbu, N, Ragunath, C, Mishra, P.J. | | Deposit date: | 2002-08-13 | | Release date: | 2002-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of a mobile loop in substrate binding and enzyme activity of
human salivary amylase.
J.Mol.Biol., 325, 2003
|
|
4OIT
 
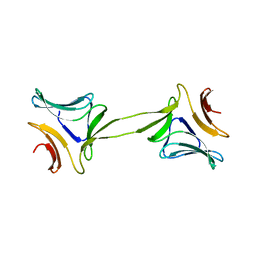 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
4WS6
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-aminouracil, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WPL
 
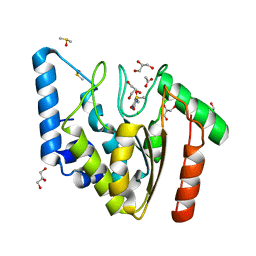 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form I | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRV
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form III | | Descriptor: | CHLORIDE ION, URACIL, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRZ
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (AB), Form I | | Descriptor: | 5-FLUOROURACIL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS2
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 6-aminouracil, Form I | | Descriptor: | 6-aminopyrimidine-2,4(3H,5H)-dione, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS7
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-chlorouracil, Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-chloropyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRX
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form V | | Descriptor: | CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS0
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (A), Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS3
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 6-aminouracil, Form IV | | Descriptor: | 6-aminopyrimidine-2,4(3H,5H)-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS8
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 2-thiouracil, Form V | | Descriptor: | 2-thioxo-2,3-dihydropyrimidin-4(1H)-one, CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS4
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-nitrouracil, Form I | | Descriptor: | 5-nitrouracil, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRU
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form II | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRW
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form IV | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
