5MM2
 
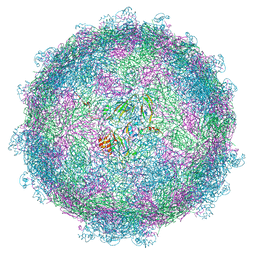 | | nora virus structure | | Descriptor: | Capsid protein VP4A, capsid protein VP4B, capsid protein VP4C | | Authors: | Laurinmaki, P, Shakeel, S, Ekstrom, J.-O, Butcher, S.J. | | Deposit date: | 2016-12-08 | | Release date: | 2017-12-20 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of Nora virus at 2.7 angstrom resolution and implications for receptor binding, capsid stability and taxonomy.
Sci Rep, 10, 2020
|
|
1L8B
 
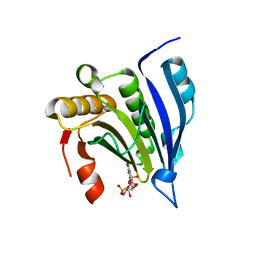 | | Cocrystal Structure of the Messenger RNA 5' Cap-binding Protein (eIF4E) bound to 7-methylGpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Niedzwiecka, A, Marcotrigiano, J, Stepinski, J, Jankowska-Anyszka, M, Wyslouch-Cieszynska, A, Dadlez, M, Gingras, A.-C, Mak, P, Darzynkiewicz, E, Sonenberg, N. | | Deposit date: | 2002-03-19 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5' cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins.
J.Mol.Biol., 319, 2002
|
|
7NYI
 
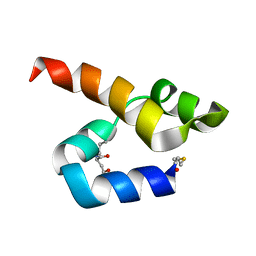 | |
5LWC
 
 | | NMR solution structure of bacteriocin BacSp222 from Staphylococcus pseudintermedius 222 | | Descriptor: | Bacteriocin BacSp222 | | Authors: | Nowakowski, M.E, Ejchart, A.O, Jaremko, L, Wladyka, B, Mak, P. | | Deposit date: | 2016-09-15 | | Release date: | 2017-10-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Spatial attributes of the four-helix bundle group of bacteriocins - The high-resolution structure of BacSp222 in solution.
Int.J.Biol.Macromol., 107, 2018
|
|
1U4D
 
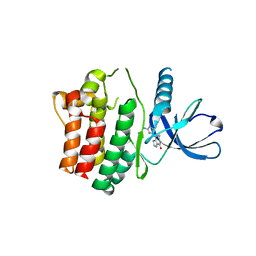 | | Structure of the ACK1 Kinase Domain bound to Debromohymenialdisine | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, DEBROMOHYMENIALDISINE | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
1U46
 
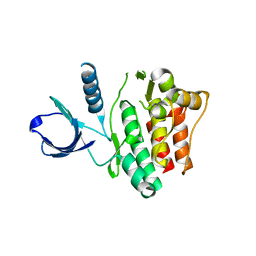 | | Crystal Structure of the Unphosphorylated Kinase Domain of the Tyrosine Kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
1U54
 
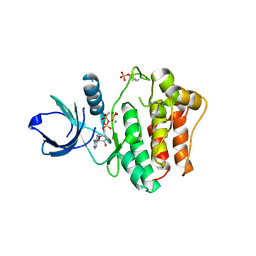 | | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the CDC42-associated Tyrosine Kinase ACK1 bound to AMP-PCP | | Descriptor: | Activated CDC42 kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
3J2J
 
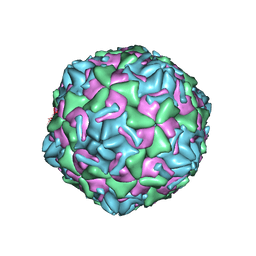 | | Empty coxsackievirus A9 capsid | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Shakeel, S, Seitsonen, J.J.T, Kajander, T, Laurinmaki, P, Hyypia, T, Susi, P, Butcher, S.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.54 Å) | | Cite: | Structural and functional analysis of coxsackievirus A9 integrin {alpha}v{beta}6 binding and uncoating.
J.Virol., 87, 2013
|
|
4BIQ
 
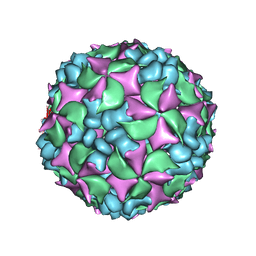 | | Homology model of coxsackievirus A7 (CAV7) empty capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
4BIP
 
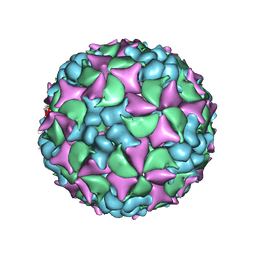 | | Homology model of coxsackievirus A7 (CAV7) full capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (8.23 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
