3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
3V69
 
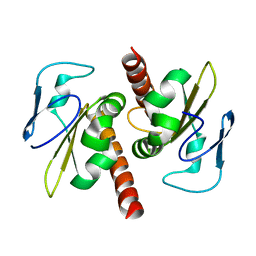 | | Filia-N crystal structure | | Descriptor: | Protein Filia | | Authors: | Wang, J, Xu, M, Zhu, K, Li, L, Liu, X. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-terminus of FILIA Forms an Atypical KH Domain with a Unique Extension Involved in Interaction with RNA.
Plos One, 7, 2012
|
|
4DVF
 
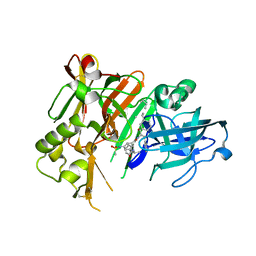 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
4DV9
 
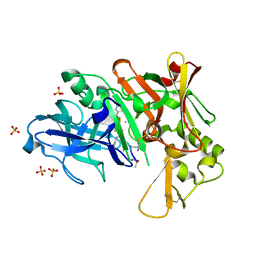 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-AMINO-3-OXOPROPYL)-2,13-DIBENZYL-12,22-DIHYDROXY-3,5,17-TRIMETHYL-8-(2-METHYLPROPYL)-4,7,10,15,18,21-HEXAOXO-19-(PROPAN-2-YL)-3,6,9,14,17,20-HEXAAZATRICOSAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE (NON-PREFERRED NAME), SULFATE ION | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
6SCX
 
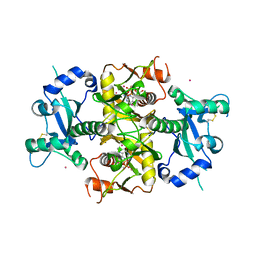 | | Crystal structure of the catalytic domain of human NUDT12 in complex with 7-methyl-guanosine-5'-triphosphate | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Peroxisomal NADH pyrophosphatase NUDT12 | | Authors: | McCarthy, A.A, Chen, K.M, Wu, H, Li, L, Homolka, D, Gos, P, Fleury-Olela, F, Pillai, R.S. | | Deposit date: | 2019-07-25 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Decapping Enzyme NUDT12 Partners with BLMH for Cytoplasmic Surveillance of NAD-Capped RNAs.
Cell Rep, 29, 2019
|
|
4FGX
 
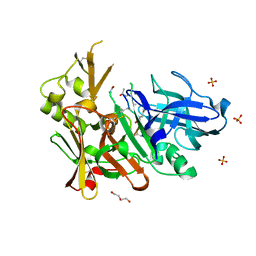 | | Crystal structure of bace1 with novel inhibitor | | Descriptor: | Beta-secretase 1, DI(HYDROXYETHYL)ETHER, Inhibitor (2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-amino-3-oxopropyl)-2,13-dibenzyl-12,22-dihydroxy-8-isobutyl-19-isopropyl-3,5,17-trimethyl-4,7,10,15,18,21-hexaoxo-3,6,9,14,17,20-hexaazatricosan-1-oic acid, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
1Q2U
 
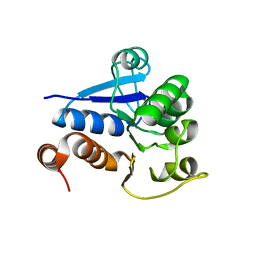 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
1Q3M
 
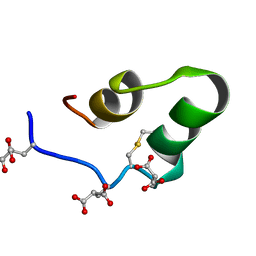 | |
6U0E
 
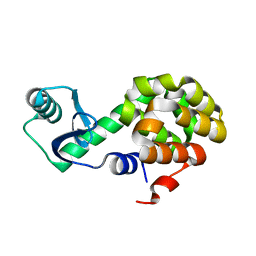 | |
6U0F
 
 | |
6U0B
 
 | |
6U0C
 
 | |
3R7W
 
 | | Crystal Structure of Gtr1p-Gtr2p complex | | Descriptor: | GTP-binding protein GTR1, GTP-binding protein GTR2, MAGNESIUM ION, ... | | Authors: | Gong, R, Li, L, Liu, Y, Wang, P, Yang, H, Wang, L, Cheng, J, Guan, K.L, Xu, Y. | | Deposit date: | 2011-03-23 | | Release date: | 2011-08-24 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation
Genes Dev., 25, 2011
|
|
4K9R
 
 | | Spore photoproduct lyase Y98F mutant | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, Spore photoproduct lyase, ... | | Authors: | Yang, L, Nelson, R.S, Benjdia, A, Lin, G, Telser, J, Stoll, S, Schlichting, I, Li, L. | | Deposit date: | 2013-04-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A radical transfer pathway in spore photoproduct lyase.
Biochemistry, 52, 2013
|
|
8H95
 
 | | Structure of mouse SCMC bound with full-length FILIA | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, L, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
1VRY
 
 | | Second and Third Transmembrane Domains of the Alpha-1 Subunit of Human Glycine Receptor | | Descriptor: | Glycine receptor alpha-1 chain | | Authors: | Ma, D, Liu, Z, Li, L, Tang, P, Xu, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Second and Third Transmembrane Domains of Human Glycine Receptor.
Biochemistry, 44, 2005
|
|
7M2P
 
 | |
7MW8
 
 | |
7N1S
 
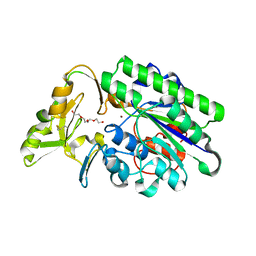 | | Crystal Structure Analysis of Xac Nucleotide Pyrophosphatase/Phosphodiesterase | | Descriptor: | Phosphodiesterase-nucleotide pyrophosphatase, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Fernandez, D, Li, L, Brown, J.A, Carozza, J.A. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ENPP1's regulation of extracellular cGAMP is a ubiquitous mechanism of attenuating STING signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3IS2
 
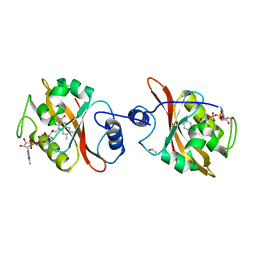 | | 2.3 Angstrom Crystal Structure of a Cys71 Sulfenic Acid form of Vivid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Vivid PAS protein VVD | | Authors: | Zoltowski, B.D, Lamb, J.S, Pabit, S.A, Li, L, Pollack, L, Crane, B.R. | | Deposit date: | 2009-08-25 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Illuminating solution responses of a LOV domain protein with photocoupled small-angle X-ray scattering.
J.Mol.Biol., 393, 2009
|
|
3HBF
 
 | | Structure of UGT78G1 complexed with myricetin and UDP | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
3HBJ
 
 | | Structure of UGT78G1 complexed with UDP | | Descriptor: | Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
4LUQ
 
 | |
4IVT
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(1H-indol-1-yl)acetamide, SULFATE ION | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
