5N4S
 
 | | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-D-tryptophan (Compound 3) | | Descriptor: | (2~{R})-3-(1~{H}-indol-3-yl)-2-[[(2~{S})-2-methyl-3-sulfanyl-propanoyl]amino]propanoic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5N58
 
 | | di-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-D-tryptophan (Compound 1) | | Descriptor: | (2~{R})-2-[(1-chloranyl-4-oxidanyl-isoquinolin-3-yl)carbonylamino]-3-(1~{H}-indol-3-yl)propanoic acid, Class B metallo-beta-lactamase, GLYCEROL, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5NAI
 
 | | mono-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-D-tryptophan (Compound 1) | | Descriptor: | (2~{R})-2-[(1-chloranyl-4-oxidanyl-isoquinolin-3-yl)carbonylamino]-3-(1~{H}-indol-3-yl)propanoic acid, Class B metallo-beta-lactamase, FLUORIDE ION, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5N55
 
 | | mono-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-L-tryptophan (Compound 2) | | Descriptor: | (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-L-tryptophan, Class B metallo-beta-lactamase, ZINC ION | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5N4T
 
 | | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-L-tryptophan (Compound 4) | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[[(2~{S})-2-methyl-3-sulfanyl-propanoyl]amino]propanoic acid, BENZOIC ACID, Beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
7BQA
 
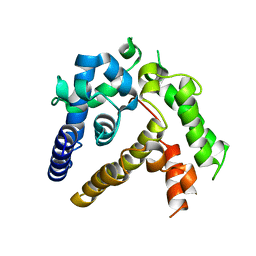 | | Crystal structure of ASFV p35 | | Descriptor: | 60 kDa polyprotein | | Authors: | Li, G.B, Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
5LCA
 
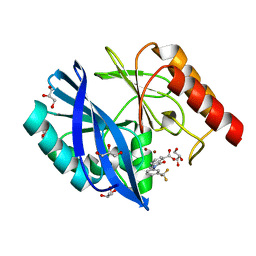 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-(3-(trifluoromethyl)phenyl)isoindoline-4-carboxylic acid (compound 17) | | Descriptor: | 3-oxidanylidene-2-[3-(trifluoromethyl)phenyl]-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LCF
 
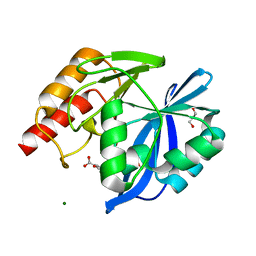 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-phenylisoindoline-4-carboxylic acid (compound 30) | | Descriptor: | 3-oxidanylidene-2-phenyl-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LE1
 
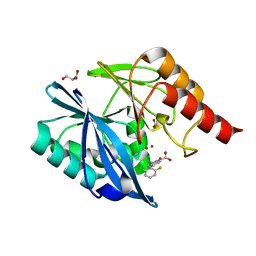 | | VIM-2 metallo-beta-lactamase in complex with 2-(2-chloro-6-fluorobenzyl)-3-oxoisoindoline-4-carboxylic acid (compound 16) | | Descriptor: | 2-[(2-chloranyl-6-fluoranyl-phenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LM6
 
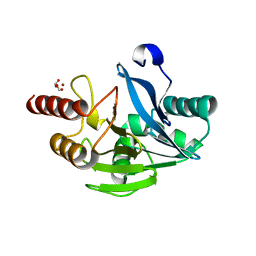 | | VIM-2 metallo-beta-lactamase in complex with 2-(3-fluoro-4-hydroxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 35) | | Descriptor: | 2-(3-fluoranyl-4-oxidanyl-phenyl)-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, Metallo-beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, Someya, H, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LCH
 
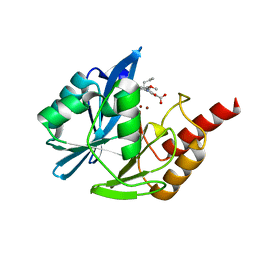 | | VIM-2 metallo-beta-lactamase in complex with (S)-1-allyl-2-(3-methoxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 42) | | Descriptor: | (1~{S})-2-(3-methoxyphenyl)-3-oxidanylidene-1-prop-2-enyl-1~{H}-isoindole-4-carboxylic acid, Metallo-beta-lactamase VIM-2, ZINC ION | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
4E2A
 
 | |
8GUE
 
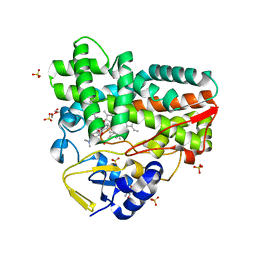 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-09-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXS
 
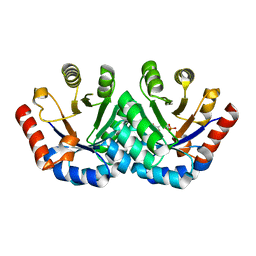 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3H6X
 
 | |
5JPU
 
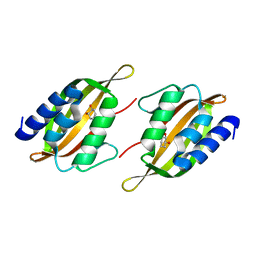 | | Structure of limonene epoxide hydrolase mutant - H-2-H5 complex with (S,S)-cyclohexane-1,2-diol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, limonene epoxide hydrolase | | Authors: | Li, G, Zhang, H, Sun, Z, Liu, X, Reetz, M.T. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
To be published
|
|
5JPP
 
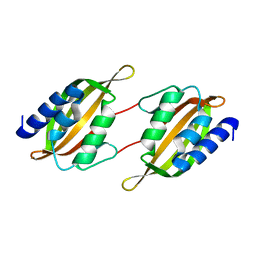 | | Structure of limonene epoxide hydrolase mutant - H-2-H5 | | Descriptor: | limonene epoxide hydrolase | | Authors: | Li, G, Zhang, H, Sun, Z, Liu, X, Reetz, M.T. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
To be published
|
|
7M4S
 
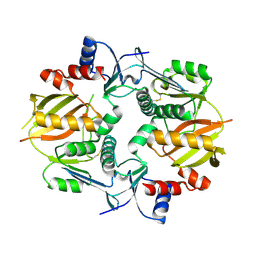 | |
2K29
 
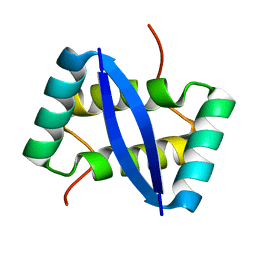 | |
2KC8
 
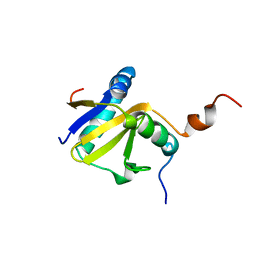 | | Structure of E. coli toxin RelE (R81A/R83A) mutant in complex with antitoxin RelBc (K47-L79) peptide | | Descriptor: | Antitoxin RelB, Toxin relE | | Authors: | Li, G, Zhang, Y, Inouye, M, Ikura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site.
J.Biol.Chem., 284, 2009
|
|
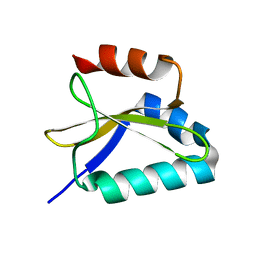
2KC9
 
 | |
7E0P
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 4-(((6-Bromo-1H-indazol-4-yl)amino)methyl)phenol (2) | | Descriptor: | 4-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]phenol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.635 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0T
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((5-bromo-1H-indazol-4-yl)amino)methyl)Cyclohexan-1-ol (36) | | Descriptor: | (1~{R},2~{S})-2-[[(5-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0S
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((6-Bromo-1H-indazol-4-yl)amino)methyl)cyclohexan-1-ol (23) | | Descriptor: | (1~{R},2~{S})-2-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
