4Z1H
 
 | | Crystal structure of human Trap1 with SMTIN-P01 | | Descriptor: | 8-[(6-iodo-1,3-benzodioxol-5-yl)sulfanyl]-9-[6-(triphenyl-lambda~5~-phosphanyl)hexyl]-9H-purin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Lee, C, Park, H.K, Ryu, J.H, Kang, B.H. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of a Mitochondria-Targeted Hsp90 Inhibitor Based on the Crystal Structures of Human TRAP1
J.Am.Chem.Soc., 137, 2015
|
|
4Z1I
 
 | | Crystal structure of human Trap1 with AMPPNP | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, MAGNESIUM ION, ... | | Authors: | Lee, C, Park, H.K, Ryu, J.H, Kang, B.H. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development of a Mitochondria-Targeted Hsp90 Inhibitor Based on the Crystal Structures of Human TRAP1
J.Am.Chem.Soc., 137, 2015
|
|
4Z1G
 
 | | Crystal structure of human Trap1 with BIIB-021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Lee, C, Park, H.K, Ryu, J.H, Kang, B.H. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-22 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Mitochondria-Targeted Hsp90 Inhibitor Based on the Crystal Structures of Human TRAP1
J.Am.Chem.Soc., 137, 2015
|
|
4Z1F
 
 | | Crystal structure of human Trap1 with PU-H71 | | Descriptor: | 8-[(6-IODO-1,3-BENZODIOXOL-5-YL)THIO]-9-[3-(ISOPROPYLAMINO)PROPYL]-9H-PURIN-6-AMINE, Heat shock protein 75 kDa, mitochondrial | | Authors: | Lee, C, Park, H.K, Ryu, J.H, Kang, B.H. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Development of a Mitochondria-Targeted Hsp90 Inhibitor Based on the Crystal Structures of Human TRAP1
J.Am.Chem.Soc., 137, 2015
|
|
1IZ3
 
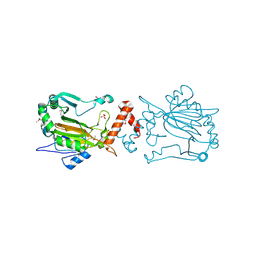 | | Dimeric structure of FIH (Factor inhibiting HIF) | | Descriptor: | FIH, SULFATE ION | | Authors: | Lee, C, Kim, S.-J, Jeong, D.-G, Lee, S.M, Ryu, S.-E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau.
J.Biol.Chem., 278, 2003
|
|
1N4M
 
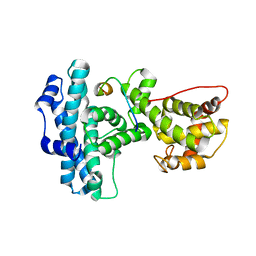 | | Structure of Rb tumor suppressor bound to the transactivation domain of E2F-2 | | Descriptor: | Retinoblastoma Pocket, Transcription factor E2F2 | | Authors: | Lee, C, Chang, J.H, Lee, H.S, Cho, Y. | | Deposit date: | 2002-10-31 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
GENES DEV., 16, 2002
|
|
2KCC
 
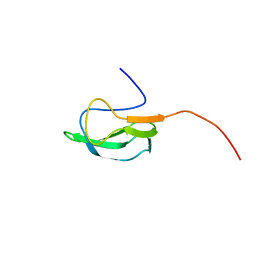 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
4BWZ
 
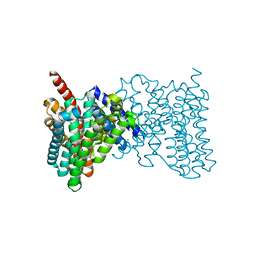 | |
5VRE
 
 | |
3L9P
 
 | |
5H20
 
 | | X-ray structure of PadR-like Transcription factor from bacteroid fragilis | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Putative PadR-family transcriptional regulatory protein, ... | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of BF2549, a PadR-like transcription factor from Bacteroides fragilis.
Biochem. Biophys. Res. Commun., 483, 2017
|
|
2JTN
 
 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | Descriptor: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | Authors: | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2007-08-03 | | Release date: | 2008-06-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
5XEF
 
 | | Crystal structure of flagellar chaperone from bacteria | | Descriptor: | Flagellar protein fliS | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the flagellar chaperone FliS from Bacillus cereus and an invariant proline critical for FliS dimerization and flagellin recognition
Biochem. Biophys. Res. Commun., 487, 2017
|
|
6ABT
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
6ABQ
 
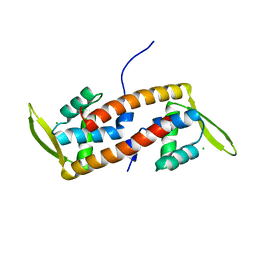 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
3MKR
 
 | |
3MKQ
 
 | |
2HIW
 
 | |
2JRL
 
 | | Solution structure of the beryllofluoride-activated NtrC4 receiver domain dimer | | Descriptor: | Transcriptional regulator (NtrC family) | | Authors: | Lee, C, Hong, E, Doucleff, M, Pelton, J.G, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Beryllofluoride-Activated NtrC4 Receiver Domain Dimer.
To be Published
|
|
2LKP
 
 | | solution structure of apo-NmtR | | Descriptor: | HTH-type transcriptional regulator NmtR | | Authors: | Lee, C, Giedroc, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mycobacterium tuberculosis NmtR in the Apo State: Insights into Ni(II)-Mediated Allostery.
Biochemistry, 51, 2012
|
|
2L14
 
 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
2L6I
 
 | |
4ZTT
 
 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | Bacterial non-heme ferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
5XQH
 
 | | Crystal structure of truncated human Rogdi | | Descriptor: | Protein rogdi homolog | | Authors: | Lee, H, Lee, C. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome
Sci Rep, 7, 2017
|
|
8J1O
 
 | | Crystal structure of HaloTag complexed with BTTA | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, N-[[1-[2-[2-(2-hexoxyethoxy)ethoxy]ethyl]-1,2,3-triazol-4-yl]methyl]-1-(1H-1,2,3-triazol-4-yl)-N-(2H-1,2,3-triazol-4-ylmethyl)methanamine, ... | | Authors: | Kim, H, Rhee, H, Lee, C. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of HaloTag complexed with BTTA
To Be Published
|
|
