7BHQ
 
 | |
7BIN
 
 | | Salmonella export gate and rod refined in focussed C1 map | | Descriptor: | Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, Flagellar basal-body rod protein FlgF, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-01-12 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7BC6
 
 | | Cryo-EM structure of the outward open proton coupled folate transporter at pH 7.5 | | Descriptor: | Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
7BC7
 
 | | Cryo-EM structure of the proton coupled folate transporter at pH 6.0 bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
6RQJ
 
 | | Structure of human complement C5 complexed with tick inhibitors OmCI, RaCI1 and CirpT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5, Complement inhibitor, ... | | Authors: | Reichhardt, M.P, Johnson, S, Lea, S.M. | | Deposit date: | 2019-05-15 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An inhibitor of complement C5 provides structural insights into activation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6S3S
 
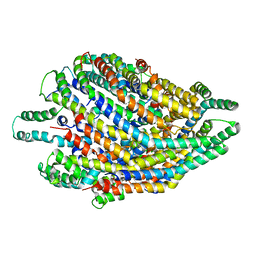 | | Structure of the FliPQR complex from the flagellar type 3 secretion system of Vibrio mimicus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Kuhlen, L, Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion.
Nat Commun, 11, 2020
|
|
6RPT
 
 | |
6S3L
 
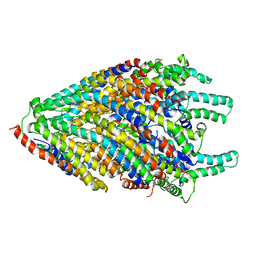 | | Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex. | | Descriptor: | Flagellar biosynthetic protein FlhB, Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, ... | | Authors: | Kuhlen, L, Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion.
Nat Commun, 11, 2020
|
|
6S3R
 
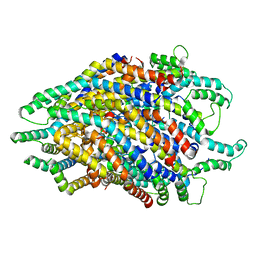 | | Structure of the FliPQR complex from the flagellar type 3 secretion system of Pseudomonas savastanoi. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Kuhlen, L, Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion.
Nat Commun, 11, 2020
|
|
6SD1
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 33-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SCN
 
 | | 33mer structure of the Salmonella flagella MS-ring protein FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-03-18 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SAN
 
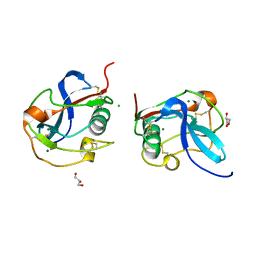 | | SALSA / DMBT1 / GP340 SRCR domain 8 soaked in calcium and magnesium | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein, GLYCEROL, ... | | Authors: | Reichhardt, M.P, Johnson, S, Loimaranta, V, Lea, S.M. | | Deposit date: | 2019-07-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
6SA5
 
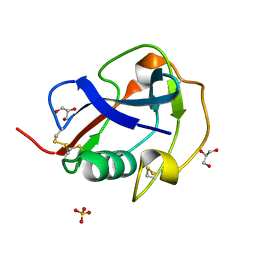 | | SALSA / DMBT1 / GP340 SRCR domain 8 | | Descriptor: | Deleted in malignant brain tumors 1 protein, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Reichhardt, M.P, Johnson, S, Loimaranta, V, Lea, S.M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
6SD2
 
 | | Structure of the RBM2inner region of the Salmonella flagella MS-ring protein FliF with 21-fold symmetry applied. | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD3
 
 | | 34mer structure of the Salmonella flagella MS-ring protein FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SA4
 
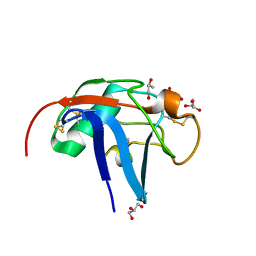 | | SALSA / DMBT1 / GP340 SRCR domain 1 | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein, GLYCEROL, ... | | Authors: | Reichhardt, M.P, Lea, S.M, Johnson, S. | | Deposit date: | 2019-07-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
7SAZ
 
 | |
7SAX
 
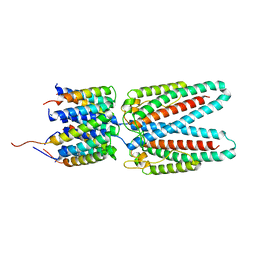 | |
7SAU
 
 | |
7SB2
 
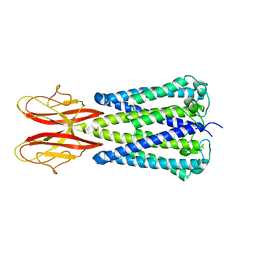 | |
7SAT
 
 | |
6IAI
 
 | |
1RXL
 
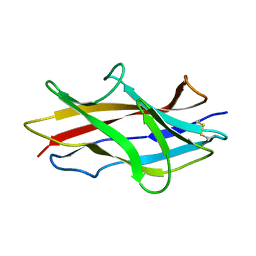 | | Solution structure of the engineered protein Afae-dsc | | Descriptor: | Afimbrial adhesin AFA-III | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, du Merle, L, Barlow, P.N, Medof, M.E, Smith, R.A, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-18 | | Release date: | 2005-01-11 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | An atomic resolution model for assembly, architecture, and function of the Dr adhesins.
Mol.Cell, 15, 2004
|
|
4B4A
 
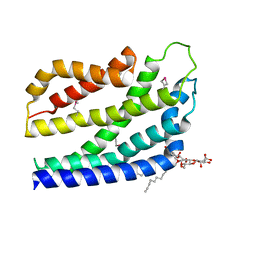 | | Structure of the TatC core of the twin arginine protein translocation system | | Descriptor: | Lauryl Maltose Neopentyl Glycol, SEC-INDEPENDENT PROTEIN TRANSLOCASE PROTEIN TATC | | Authors: | Rollauer, S.E, Tarry, M.J, Jaaskelainen, M, Graham, J.E, Jaeger, F, Krehenbrink, M, Roversi, P, McDowell, M.A, Stansfeld, P.J, Johnson, S, Liu, S.M, Lukey, M.J, Marcoux, J, Robinson, C.V, Sansom, M.S, Palmer, T, Hogbom, M, Berks, B.C, Lea, S.M. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-05 | | Last modified: | 2012-12-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Tatc Core of the Twin-Arginine Protein Transport System.
Nature, 49, 2012
|
|
1UPN
 
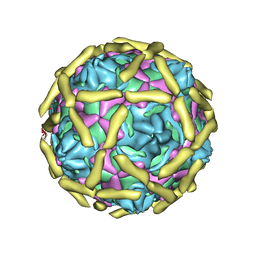 | | COMPLEX OF ECHOVIRUS TYPE 12 WITH DOMAINS 3 AND 4 OF ITS RECEPTOR DECAY ACCELERATING FACTOR (CD55) BY CRYO ELECTRON MICROSCOPY AT 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Bhella, D, Goodfellow, I.G, Roversi, P, Pettigrew, D, Chaudry, Y, Evans, D.J, Lea, S.M. | | Deposit date: | 2003-10-08 | | Release date: | 2004-01-07 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The Structure of Echovirus Type 12 Bound to a Two-Domain Fragment of its Cellular Attachment Protein Decay-Accelerating Factor (Cd 55)
J.Biol.Chem., 279, 2004
|
|
