7B5E
 
 | | Structure of calcium-bound mTMEM16A(ac)-I551A chloride channel at 4.1 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7B5D
 
 | |
7B5C
 
 | | Structure of calcium-bound mTMEM16A(ac) chloride channel at 3.7 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7ZK3
 
 | | Structure of 1PBC- and calcium-bound mTMEM16A(ac) chloride channel at 2.85 A resolution | | Descriptor: | 1-Hydroxy-3-(trifluoromethyl)pyrido[1,2-a]benzimidazole-4-carbonitrile, Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rutz, S, Dutzler, R. | | Deposit date: | 2022-04-12 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Inhibition mechanism of the chloride channel TMEM16A by the pore blocker 1PBC.
Nat Commun, 13, 2022
|
|
8QZC
 
 | |
4OSG
 
 | | Klebsiella pneumoniae complexed with NADPH and 6-ethyl-5-[(3R)-3-[3-methoxyl-5-(pyridine-4-yl)phenyl]but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP1006) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Klebsiella pneumoniae Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal Features for Potency and Selectivity.
Antimicrob.Agents Chemother., 58, 2014
|
|
4OR7
 
 | |
4KEB
 
 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5-(isoquin-5-yl)phenyl]but-1-yn-1-yl}6-ethylpyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{(3S)-3-[3-(isoquinolin-5-yl)-5-methoxyphenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
4KAK
 
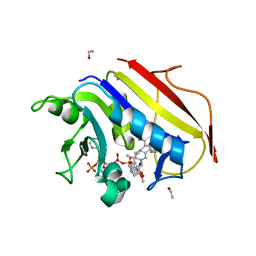 | | Crystal structure of human dihydrofolate reductase complexed with NADPH and 6-ethyl-5-[(3S)-3-[3-methoxy-5-(pyridine-4-yl)phenyl]but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP1006) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-22 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
4KBN
 
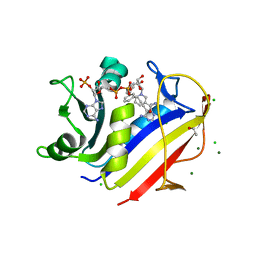 | | human dihydrofolate reductase complexed with NADPH and 5-{3-[3-(3,5-pyrimidine)]-phenyl-prop-1-yn-1-yl}-6-ethyl-pyrimidine-2,4diamine | | Descriptor: | 6-ethyl-5-{3-[3-(pyrimidin-5-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
4KD7
 
 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5(pyridine-4-yl)phenyl]prop-1-yn-1-yl}-6-ethyl-pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[3-methoxy-5-(pyridin-4-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-24 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.715 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
4KFJ
 
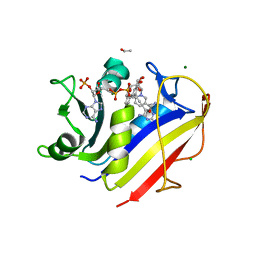 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5-(isoquin-5-yl)phenyl]prop-1-yn-1-yl}6-ethylprimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[3-(isoquinolin-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
5OYG
 
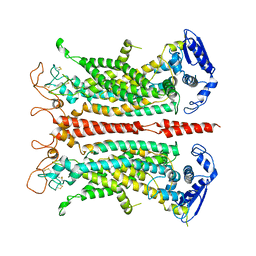 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
5OYB
 
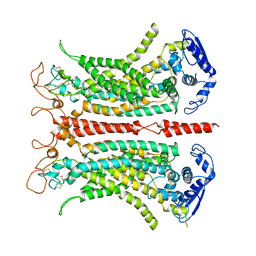 | | Structure of calcium-bound mTMEM16A chloride channel at 3.75 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2018-02-28 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
6G9O
 
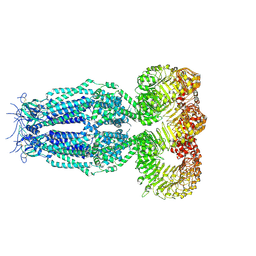 | | Structure of full-length homomeric mLRRC8A volume-regulated anion channel at 4.25 A resolution | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Sawicka, M, Deneka, D, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-04-11 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
6G8Z
 
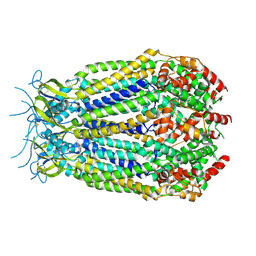 | | Structure of the pore domain of homomeric mLRRC8A volume-regulated anion channel at 3.66 A resolution | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Sawicka, M, Deneka, D, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
6FNW
 
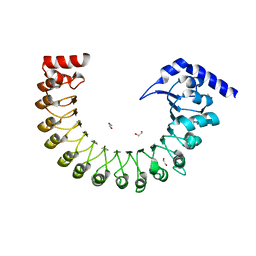 | | Structure of a volume-regulated anion channel of the LRRC8 family | | Descriptor: | 1,2-ETHANEDIOL, Volume-regulated anion channel subunit LRRC8A | | Authors: | Deneka, D, Sawicka, M, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-02-05 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
6G9L
 
 | | Structure of homomeric mLRRC8A volume-regulated anion channel at 5.01 A resolution | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Sawicka, M, Deneka, D, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-04-11 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
