3PTE
 
 | |
9LYZ
 
 | |
7NVM
 
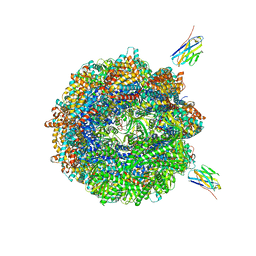 | | Human TRiC complex in closed state with nanobody Nb18, actin and PhLP2A bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Actin, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVL
 
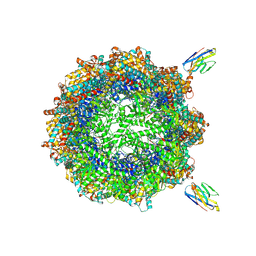 | | Human TRiC complex in closed state with nanobody bound (Consensus Map) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVO
 
 | | Human TRiC complex in open state with nanobody bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVN
 
 | | Human TRiC complex in closed state with nanobody and tubulin bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RTP
 
 | |
1BM7
 
 | |
1BMZ
 
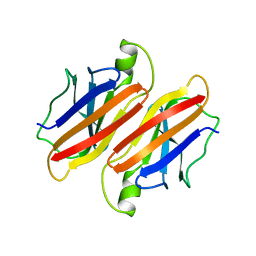 | |
1IKI
 
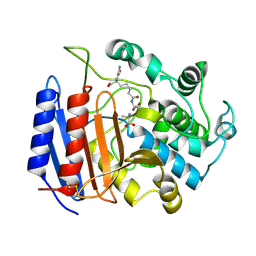 | | COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH THE PRODUCTS OF A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANINE, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1IKG
 
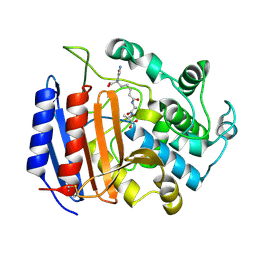 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
8VE0
 
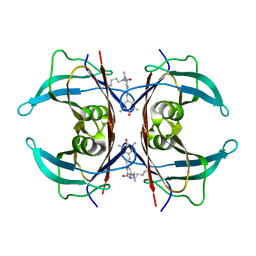 | | Human transthyretin covalently modified with A2-derived stilbene in the compressed conformation | | Descriptor: | 3-(dimethylamino)-5-[(E)-2-(4-hydroxy-3,5-dimethylphenyl)ethenyl]benzoic acid, Transthyretin | | Authors: | Basanta, B, Nugroho, K, Yan, N, Kline, G.M, Tsai, F.J, Wu, M, Kelly, J.W, Lander, G.C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The conformational landscape of human transthyretin revealed by cryo-EM
To Be Published
|
|
5HZQ
 
 | | Crystal structure of cellular retinoic acid binding protein 2 (CRABP2)-aryl fluorosulfate covalent conjugate | | Descriptor: | 4'-[(3,6,9,12-tetraoxapentadec-14-yn-1-yl)oxy][1,1'-biphenyl]-4-yl sulfurofluoridate, Cellular retinoic acid-binding protein 2, GLYCEROL | | Authors: | Chen, W, Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2016-02-02 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue.
J.Am.Chem.Soc., 138, 2016
|
|
1MPL
 
 | | CRYSTAL STRUCTURE OF PHOSPHONATE-INHIBITED D-ALA-D-ALA PEPTIDASE REVEALS AN ANALOG OF A TETRAHEDRAL TRANSITION STATE | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, GLYCYL-L-A-AMINOPIMELYL-E-(D-2-AMINOETHYL)PHOSPHONATE | | Authors: | Silvaggi, N.R, Anderson, J.W, Brinsmade, S.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2002-09-12 | | Release date: | 2003-02-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Crystal Structure of Phosphonate-Inhibited
d-Ala-d-Ala Peptidase Reveals an Analogue of a Tetrahedral Transition State.
Biochemistry, 42, 2003
|
|
1HVB
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES R61 DD-PEPTIDASE COMPLEXED WITH A NOVEL CEPHALOSPORIN ANALOG OF CELL WALL PEPTIDOGLYCAN | | Descriptor: | 5-{3-(S)-(4-(R)-ACETYLAMINO-4-CARBOXY-BUTYRYLAMINO)-3-[1-(R)-(1-(R)-CARBOXY-ETHYLCARBAMOYL)-ETHYLCARBAMOYL]-PROPYL}-2-( CARBOXY-PHENYLACETYLAMINO-METHYL)-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE | | Authors: | McDonough, M.A, Lee, W, Silvaggi, N.R, Mobashery, S, Kelly, J.A. | | Deposit date: | 2001-01-08 | | Release date: | 2001-02-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A 1.2-A snapshot of the final step of bacterial cell wall biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5L9A
 
 | | L-threonine dehydrogenase from trypanosoma brucei. | | Descriptor: | ACETATE ION, L-threonine 3-dehydrogenase | | Authors: | Erskine, P.T, Cooper, J.B, Adjogatse, E, Kelly, J, Wood, S.P. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6I2K
 
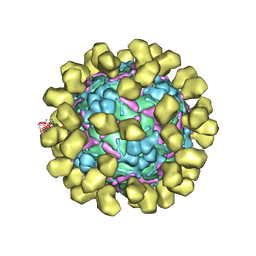 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
8FO3
 
 | |
8FO5
 
 | |
8FO4
 
 | |
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
5UI4
 
 | | Structure of NME1 covalently conjugated to imidazole fluorosulfate | | Descriptor: | 4-[4-(3-methoxyphenyl)-1-(prop-2-yn-1-yl)-1H-imidazol-5-yl]phenyl sulfurofluoridate, Nucleoside diphosphate kinase A | | Authors: | Mortenson, D.E, Brighty, G.J, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
5UEH
 
 | | Structure of GSTO1 covalently conjugated to quinolinic acid fluorosulfate | | Descriptor: | 2-(4-chlorophenyl)-6-[(fluorosulfonyl)oxy]quinoline-4-carboxylic acid, GLYCEROL, Glutathione S-transferase omega-1, ... | | Authors: | Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
1PWD
 
 | | Covalent acyl enzyme complex of the Streptomyces R61 DD-peptidase with cephalosporin C | | Descriptor: | (2R)-5-(acetyloxymethyl)-2-[(1R)-1-[[(5R)-5-azanyl-6-oxidanyl-6-oxidanylidene-hexanoyl]amino]-2-oxidanylidene-ethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase precursor | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1PW1
 
 | | Non-Covalent Complex Of Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2S,5R,6R)-6-{[(6R)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, FORMYL GROUP, ... | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
