7CM8
 
 | | High resolution crystal structure of M92A mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase, GLYCEROL, SODIUM ION | | Authors: | Kaushik, A, Rahisuddin, R, Saini, N, Kumaran, S. | | Deposit date: | 2020-07-25 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of selective substrate engagement and inhibitor disengagement of cysteine synthase.
J.Biol.Chem., 296, 2020
|
|
4ORE
 
 | |
5DBH
 
 | |
4RUH
 
 | | Crystal structure of Human Carnosinase-2 (CN2) in complex with inhibitor, Bestatin at 2.25 A | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Cytosolic non-specific dipeptidase, GLYCEROL, ... | | Authors: | pandya, V, Kaushik, A, Singh, A.K, Singh, R.P, Kumaran, S. | | Deposit date: | 2014-11-19 | | Release date: | 2015-11-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Human Carnosinase-2 (CN2) in complex with inhibitor, Bestatin at 2.25 A
TO BE PUBLISHED
|
|
4ZU1
 
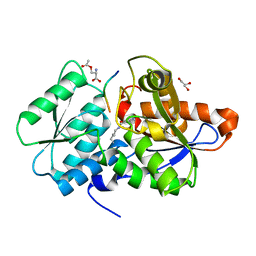 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus influenzae in complex with O-acetyl serine and peptide inhibitor | | Descriptor: | C-terminal peptide from Serine acetyltransferase, Cysteine synthase, GLYCEROL, ... | | Authors: | Ekka, M.K, Singh, A.K, Kaushik, A, Kumaran, S. | | Deposit date: | 2015-05-15 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus inuen-zae in complex with O-acetyl serine and peptide inhibitor
To Be Published
|
|
4ZU6
 
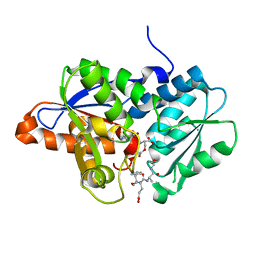 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus influenzae in complex with pre-reactive o-acetyl serine, alpha-aminoacrylate reaction intermediate and Peptide inhibitor at the resolution of 2.25A | | Descriptor: | C-terminal peptide from Serine acetyltransferase, Cysteine synthase, O-ACETYLSERINE, ... | | Authors: | Ekka, M.K, Singh, A.K, Kaushik, A, Kumaran, S. | | Deposit date: | 2015-05-15 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus inuen-zae in complex with pre-reactive o-acetyl serine and peptide inhibitor
To Be Published
|
|
7MD9
 
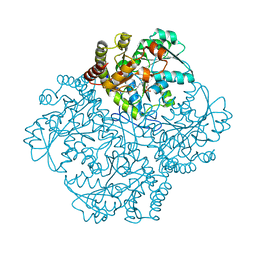 | |
7MD6
 
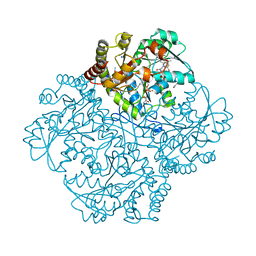 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant co-crystallized with NL1 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CITRATE ANION, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MDB
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103A mutant co-crystallized with NL2 | | Descriptor: | 5-[(6-bromo-1H-indol-1-yl)methyl]-2-methylfuran-3-carboxylic acid, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CACODYLATE ION, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MD1
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MD8
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant co-crystallized with NL2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MDA
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103A mutant co-crystallized with NL1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
6VCQ
 
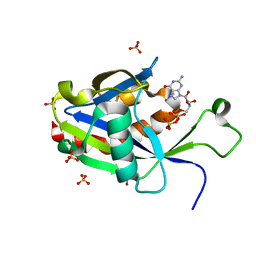 | | Crystal structure of E.coli RppH in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCN
 
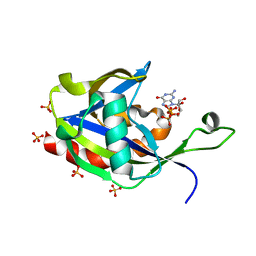 | | Crystal structure of E.coli RppH in complex with ppcpG | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCR
 
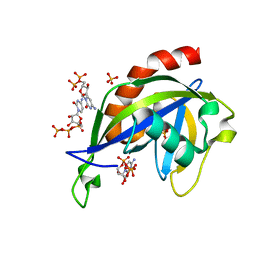 | | Crystal structure of E.coli RppH in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, PYROPHOSPHATE, RNA pyrophosphohydrolase, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCK
 
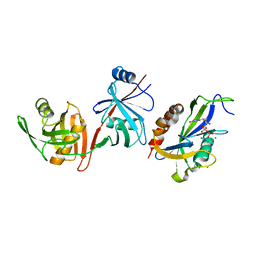 | | Crystal structure of E.coli RppH-DapF in complex with GDP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCP
 
 | | Crystal structure of E.coli RppH in complex with UTP | | Descriptor: | RNA pyrophosphohydrolase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCM
 
 | | Crystal structure of E.coli RppH-DapF in complex with GTP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCO
 
 | | Crystal structure of E.coli RppH in complex with ppcpA | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, RNA pyrophosphohydrolase | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCL
 
 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6AIF
 
 | |
4NU8
 
 | |
4LI3
 
 | |
7MCB
 
 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase Holoenzyme | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MCQ
 
 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, AOAA-bound enzyme in dimeric form | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
