5FHM
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-(aminomethyl)benzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at resolution 1.55 A resolution | | Descriptor: | (2~{S})-3-[5-[2-[[3-(aminomethyl)phenyl]methyl]-1,2,3,4-tetrazol-5-yl]-3-oxidanyl-1,2-oxazol-4-yl]-2-azanyl-propanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Al-musaed, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
1FY1
 
 | | [R23S,F25E]HBP, A MUTANT OF HUMAN HEPARIN BINDING PROTEIN (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHANOL, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
1FY3
 
 | | [G175Q]HBP, A mutant of human heparin binding protein (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
2AIX
 
 | |
1TN3
 
 | | THE C-TYPE LECTIN CARBOHYDRATE RECOGNITION DOMAIN OF HUMAN TETRANECTIN | | Descriptor: | CALCIUM ION, ETHANOL, SULFATE ION, ... | | Authors: | Kastrup, J.S, Nielsen, B.B, Rasmussen, H, Holtet, T.L, Graversen, J.H, Etzerodt, M, Thoegersen, H.C, Larsen, I.K. | | Deposit date: | 1997-11-06 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-type lectin carbohydrate recognition domain of human tetranectin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6FZ4
 
 | | Structure of GluK1 ligand-binding domain in complex with N-(7-fluoro-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)-2-hydroxybenzamide at 1.85 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Mollerud, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N1-Substituted Quinoxaline-2,3-diones as Kainate Receptor Antagonists: X-ray Crystallography, Structure-Affinity Relationships, and in Vitro Pharmacology.
Acs Chem Neurosci, 10, 2019
|
|
3BBR
 
 | |
1BFF
 
 | | THE 154 AMINO ACID FORM OF HUMAN BASIC FIBROBLAST GROWTH FACTOR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR, BETA-MERCAPTOETHANOL, PHOSPHATE ION | | Authors: | Kastrup, J.S, Eriksson, E.S. | | Deposit date: | 1996-12-06 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the 154-amino-acid form of recombinant human basic fibroblast growth factor. comparison with the truncated 146-amino-acid form.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1WVJ
 
 | | Exploring the GluR2 ligand-binding core in complex with the bicyclic AMPA analogue (S)-4-AHCP | | Descriptor: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, GLYCEROL, SULFATE ION, ... | | Authors: | Nielsen, B.B, Pickering, D.S, Greenwood, J.R, Brehm, L, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the GluR2 ligand-binding core in complex with the bicyclical AMPA analogue (S)-4-AHCP
FEBS J., 272, 2005
|
|
5FHO
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-chlorobenzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at 2.3 A resolution | | Descriptor: | (1S)-1-carboxy-2-(5-{2-[(3-chlorophenyl)methyl]-2H-tetrazol-5-yl}-3-oxo-2,3-dihydro-1,2-oxazol-4-yl)ethan-1-aminium, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
5FHN
 
 | |
7Z2R
 
 | | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics | | Descriptor: | Glutamate receptor ionotropic, delta-1, SULFATE ION | | Authors: | Masternak, M, Laulumaa, S, Kastrup, J.S. | | Deposit date: | 2022-02-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics.
Febs J., 290, 2023
|
|
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
1MS7
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.97 A resolution, Crystallization in the presence of zinc acetate | | Descriptor: | (S)-2-AMINO-3-(3-HYDROXY-ISOXAZOL-4-YL)PROPIONIC ACID, Glutamate receptor subunit 2, ZINC ION | | Authors: | Kasper, C, Lunn, M.-L, Liljefors, T, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2002-09-19 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | GluR2 ligand-binding core complexes: importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists
FEBS Lett., 531, 2002
|
|
4N07
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold.
J.Med.Chem., 56, 2013
|
|
2FPE
 
 | | Conserved dimerization of the ib1 src-homology 3 domain | | Descriptor: | C-jun-amino-terminal kinase interacting protein 1, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guenat, S, Dar, I, Bonny, C, Kastrup, J.S, Gajhede, M, Kristensen, O. | | Deposit date: | 2006-01-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unique set of SH3-SH3 interactions controls IB1 homodimerization
Embo J., 25, 2006
|
|
3C9N
 
 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
3U8L
 
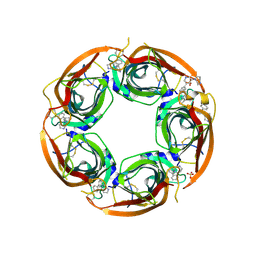 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3570 (1-(5-phenylpyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-phenylpyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
8BST
 
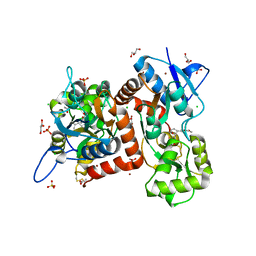 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate at 2.7A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
3U8K
 
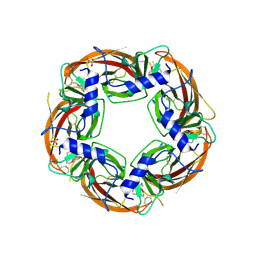 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3573 (1-(5-ethoxypyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
3U8N
 
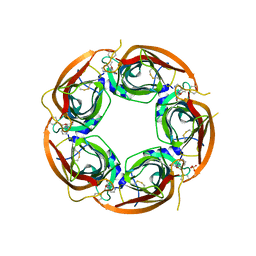 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3950 (1-(6-bromo-5-ethoxypyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(6-bromo-5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
8BSU
 
 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate and the positive allosteric modulator BPAM344 at 2.9A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Positive allosteric modulation of homomeric kainate receptors GluK1-3
To Be Published
|
|
4U4X
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM37 at 1.56 A resolution. | | Descriptor: | 4-ethyl-3,4-dihydro-2H-pyrido[3,2-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Positive Allosteric Modulators of 2-Amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionic Acid Receptors Belonging to 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-pyridothiadiazine Dioxides and Diversely Chloro-Substituted 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides.
J.Med.Chem., 57, 2014
|
|
3TZA
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid at 1.9A resolution | | Descriptor: | (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid, Glutamate receptor 2,Glutamate receptor 2, SULFATE ION | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new phenylalanine derivative acts as an antagonist at the AMPA receptor GluA2 and introduces partial domain closure: synthesis, resolution, pharmacology, and crystal structure
J.Med.Chem., 54, 2011
|
|
