1ARK
 
 | |
2JHO
 
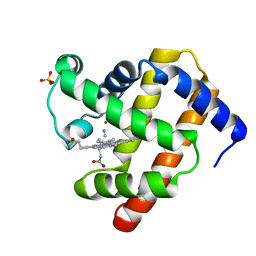 | | Cyanomet Sperm Whale Myoglobin at 1.4A resolution | | Descriptor: | CYANIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Arcovito, A, Benfatto, M, Cianci, M, Hasnain, S.S, Nienhaus, K, Nienhaus, G.U, Savino, C, Strange, R.W, Vallone, B, Della Longa, S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure analysis of a metalloprotein with enhanced active-site resolution using in situ x-ray absorption near edge structure spectroscopy.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
2VRY
 
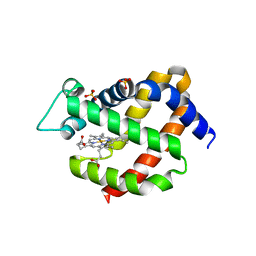 | | Mouse Neuroglobin with heme iron in the reduced ferrous state | | Descriptor: | NEUROGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Arcovito, A, Moschetti, T, D'Angelo, P, Mancini, G, Vallone, B, Brunori, M, Della Longa, S. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An X-Ray Diffraction and X-Ray Absorption Spectroscopy Joint Study of Neuroglobin.
Arch.Biochem.Biophys., 475, 2008
|
|
1NEB
 
 | |
4Y6L
 
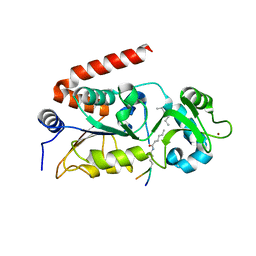 | | Human SIRT2 in complex with myristoylated peptide (H3K9myr) | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, peptide THR-ALA-ARG-MYK-SER-THR-GLY | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
4Y6Q
 
 | | Human SIRT2 in complex with 2-O-myristoyl-ADP-ribose | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, [(2S,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] tetradecanoate | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
4Y6O
 
 | |
7MEQ
 
 | | Crystal structure of human TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Transmembrane protease serine 2, ... | | Authors: | Fraser, B, Beldar, S, Hutchinson, A, Li, Y, Seitova, A, Edwards, A.M, Benard, F, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Nat.Chem.Biol., 18, 2022
|
|
5K47
 
 | | CryoEM structure of the human Polycystin-2/PKD2 TRP channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Pike, A.C.W, Grieben, M, Shintre, C.A, Tessitore, A, Shrestha, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-24 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2).
Nat. Struct. Mol. Biol., 24, 2017
|
|
7YYK
 
 | | Crystal structure of the O-fucosylated form of TSRs1-3 from the human thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1, alpha-L-fucopyranose | | Authors: | Berardinelli, S.J, Eletsky, A, Valero-Gonzalez, J, Ito, A, Manjunath, R, Hurtado-Guerrero, R, Prestegard, J.R, Woods, R.J, Haltiwanger, R.S. | | Deposit date: | 2022-02-18 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | O-fucosylation stabilizes the TSR3 motif in thrombospondin-1 by interacting with nearby amino acids and protecting a disulfide bond.
J.Biol.Chem., 298, 2022
|
|
5Y4H
 
 | | Human SIRT3 in complex with halistanol sulfate | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, THr-Arg-Ser-GLY-ALY-VAL-MET-ARG-ARG-LEU-ARG-ARG, ... | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Halistanol sulfates I and J, new SIRT1-3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria
J. Antibiot., 71, 2018
|
|
5Y0Z
 
 | | Human SIRT2 in complex with a specific inhibitor, NPD11033 | | Descriptor: | (1~{R},9~{S})-11-[(2~{R})-3-[2,4-bis(2-methylbutan-2-yl)phenoxy]-2-oxidanyl-propyl]-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-07-19 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a novel small molecule that inhibits deacetylase but not defatty-acylase reaction catalysed by SIRT2.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
5AYF
 
 | | Crystal structure of SET7/9 in complex with cyproheptadine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dibenzo[1,2-a:2',1'-d][7]annulen-11-ylidene)-1-methyl-piperidine, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Niwa, H, Handa, N, Takemoto, Y, Ito, A, Tomabechi, Y, Umehara, T, Shirouzu, M, Yoshida, M, Yokoyama, S. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription
J.Med.Chem., 59, 2016
|
|
6CVZ
 
 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
3KS9
 
 | | Metabotropic glutamate receptor mGluR1 complexed with LY341495 antagonist | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Dobrovetsky, E, Khutoreskaya, G, Seitova, A, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-20 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metabotropic glutamate receptor mglur1 complexed with LY341495 antagonist
To be Published
|
|
1O5W
 
 | | The structure basis of specific recognitions for substrates and inhibitors of rat monoamine oxidase A | | Descriptor: | Amine oxidase [flavin-containing] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | Ma, J, Yoshimura, M, Yamashita, E, Nakagawa, A, Ito, A, Tsukihara, T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of rat monoamine oxidase a and its specific recognitions for substrates and inhibitors.
J.Mol.Biol., 338, 2004
|
|
8W3V
 
 | | Crystal structure of human WDR41 | | Descriptor: | WD repeat-containing protein 41 | | Authors: | Hutchinson, A, Dong, A, Li, Y, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human WDR41
To be published
|
|
8V04
 
 | | High resolution TMPRSS2 structure following acylation by nafamostat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Levon, H, Arrowsmith, C. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High resolution TMPRSS2 structure following acylation by nafamostat
To Be Published
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
8VIS
 
 | | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Kenney, T, Li, Y.Y, Seitova, A, Li, Y, Hejazi, Z, Edwards, A, Benard, F, Arrowsmith, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide
To Be Published
|
|
8V1F
 
 | | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Halabelian, L, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol
To Be Published
|
|
1HR7
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR6
 
 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
7EDC
 
 | | Crystal structure of mutant tRNA [Gm18] methyltransferase TrmH (E107G) in complex with S-adenosyl-L-methionine from Escherichia coli | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (guanosine(18)-2'-O)-methyltransferase | | Authors: | Kono, Y, Ito, A, Okamoto, A, Yamagami, R, Hirata, A, Hori, H. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Unique substrate specificity of type II tRNA Gm18 methyltransferase from Escherichia coli
To Be Published
|
|
