3PMZ
 
 | | Crystal Structure of the Complex of Acetylcholine Binding Protein and d-tubocurarine | | Descriptor: | (1beta,1'alpha)-7',12'-dihydroxy-6,6'-dimethoxy-2,2',2'-trimethyltubocuraran-2'-ium, MAGNESIUM ION, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Harel, M, Yamauchi, J.G, Radic, Z, Hansen, S, Huxford, T, Taylor, P.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Curare Alkaloids: Analyzing the Poses of Complexes with the Acetylcholine Binding Protein in Relation to Structure and Binding Energies
To be Published
|
|
6NL7
 
 | | Crystal structure of B1 immunoglobulin-binding domain of Streptococcal Protein G (T16F, T18A, V21H, T25H, K28Y, V29I, K31R, Q32A, Y33L, N35K, D36A, N37Q) | | Descriptor: | ACETATE ION, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
1K3Z
 
 | | X-ray crystal structure of the IkBb/NF-kB p65 homodimer complex | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Shiva, M, Huang, D.B, Chen, Y, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
1OY3
 
 | | CRYSTAL STRUCTURE OF AN IKBBETA/NF-KB P65 HOMODIMER COMPLEX | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Malek, S, Huang, D.B, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
2BYS
 
 | | CRYSTAL STRUCTURE OF ACHBP FROM APLYSIA CALIFORNICA IN complex with lobeline | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN, LOBELINE | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2BYP
 
 | | Crystal structure of Aplysia californica AChBP in complex with alpha- conotoxin ImI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2BYR
 
 | | CRYSTAL STRUCTURE OF ACHBP FROM APLYSIA CALIFORNICA in complex with methyllycaconitine | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN, METHYLLYCACONITINE | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
6NL9
 
 | | Crystal structure of de novo designed metal-controlled dimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, T16L, V29H, Y33H, N37L)-apo | | Descriptor: | Immunoglobulin G-binding protein G, MAGNESIUM ION, SODIUM ION | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
6NLA
 
 | | Crystal structure of de novo designed metal-controlled dimer of B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, E15V, T16L, T18I, V29H, Y33H, N37L)-zinc | | Descriptor: | CHLORIDE ION, GLYCEROL, Immunoglobulin G-binding protein G, ... | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
6NLB
 
 | | Crystal structure of de novo designed metal-controlled dimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, E15V, T16L, T18I, V29H, Y33H, N37L)-apo | | Descriptor: | Immunoglobulin G-binding protein G, MAGNESIUM ION | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
6NL8
 
 | | Crystal structure of de novo designed metal-controlled dimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, T16L, V29H, Y33H, N37L)-zinc | | Descriptor: | CHLORIDE ION, Immunoglobulin G-binding protein G, ZINC ION | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
3NOW
 
 | | UNC-45 from Drosophila melanogaster | | Descriptor: | UNC-45 protein, SD10334p | | Authors: | Lee, C.F, Hauenstein, A.V, Fleming, J.K, Gasper, W.C, Engelke, V, Banumathi, S, Bernstein, S.I, Huxford, T. | | Deposit date: | 2010-06-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | X-ray Crystal Structure of the UCS Domain-Containing UNC-45 Myosin Chaperone from Drosophila melanogaster.
Structure, 19, 2011
|
|
2BYN
 
 | | Crystal structure of apo AChBP from Aplysia californica | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PENTAETHYLENE GLYCOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2BYQ
 
 | | Crystal structure of Aplysia californica AChBP in complex with epibatidine | | Descriptor: | EPIBATIDINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
3QCT
 
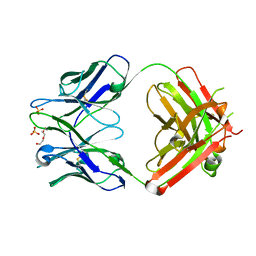 | | Crystal structure of the humanized apo LT3015 anti-lysophosphatidic acid antibody Fab fragment | | Descriptor: | LT3015 antibody Fab fragment, heavy chain, light chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1493 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
3QCU
 
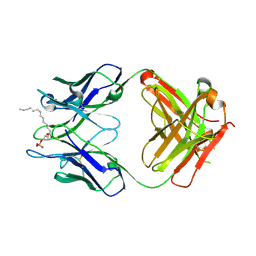 | | Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (14:0) | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, LT3015 antibody Fab fragment, heavy chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
3QCV
 
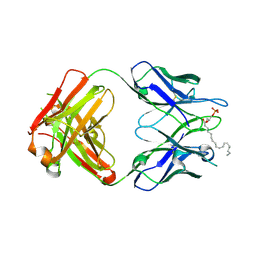 | | Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (18:2) | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9Z,12Z)-octadeca-9,12-dienoate, LT3015 antibody Fab fragment, heavy chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
4E3C
 
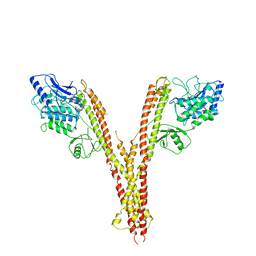 | | X-ray crystal structure of human IKK2 in an active conformation | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta | | Authors: | Polley, S, Huang, D.B, Hauenstein, A.V, Ghosh, G, Huxford, T. | | Deposit date: | 2012-03-09 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | X-ray crystal structure of human IKK2 in an active conformation
Plos.Biol., 2013
|
|
