1FKQ
 
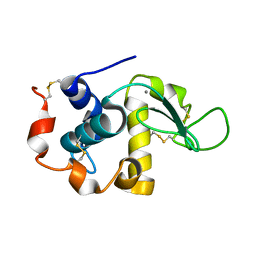 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN T29V | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 2000-08-10 | | Release date: | 2001-02-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of Thr29 to the thermodynamic stability of goat alpha-lactalbumin as determined by experimental and theoretical approaches.
Proteins, 45, 2001
|
|
1FKV
 
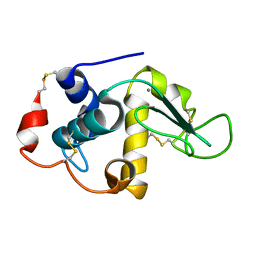 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN T29I | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 2000-08-10 | | Release date: | 2001-02-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of Thr29 to the thermodynamic stability of goat alpha-lactalbumin as determined by experimental and theoretical approaches.
Proteins, 45, 2001
|
|
2GI7
 
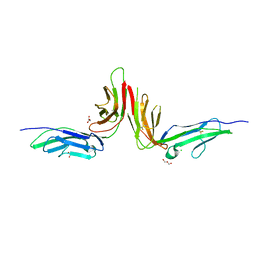 | | Crystal structure of human platelet Glycoprotein VI (GPVI) | | Descriptor: | CHLORIDE ION, GLYCEROL, GPVI protein, ... | | Authors: | Horii, K, Kahn, M.L, Herr, A.B. | | Deposit date: | 2006-03-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI.
Blood, 108, 2006
|
|
1HMK
 
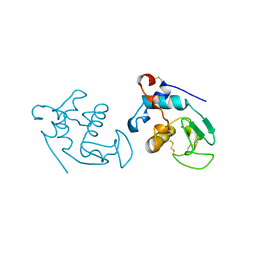 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
1UKM
 
 | | Crystal structure of EMS16, an Antagonist of collagen receptor integrin alpha2beta1 (GPIa/IIa) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EMS16 A chain, ... | | Authors: | Horii, K, Okuda, D, Morita, T, Mizuno, H. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of EMS16, an Antagonist of collagen receptor (GPIa/IIa) from the venom of Echis multisquamatus
Biochemistry, 42, 2003
|
|
1V7P
 
 | | Structure of EMS16-alpha2-I domain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EMS16 A chain, ... | | Authors: | Horii, K, Okuda, D, Morita, T, Mizuno, H. | | Deposit date: | 2003-12-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of EMS16 in complex with the integrin alpha2-I domain
J.Mol.Biol., 341, 2004
|
|
3WOC
 
 | | Crystal structure of a prostate-specific WGA16 glycoprotein lectin, form II | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Garenaux, E, Kanagawa, M, Tsuchiyama, T, Hori, K, Kanazawa, T, Goshima, A, Chiba, M, Yasue, H, Ikeda, A, Yamaguchi, Y, Sato, C, Kitajima, K. | | Deposit date: | 2013-12-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery, Primary, and Crystal Structures and Capacitation-related Properties of a Prostate-derived Heparin-binding Protein WGA16 from Boar Sperm
J.Biol.Chem., 290, 2015
|
|
3WP8
 
 | | Acinetobacter sp. Tol 5 AtaA C-terminal Ylhead fused to GCN4 adaptors (Chead) | | Descriptor: | Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPO
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1i) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-14 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPA
 
 | | Acinetobacter sp. Tol 5 AtaA C-terminal stalk_FL fused to GCN4 adaptors (CstalkFL) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPP
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1iii) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPR
 
 | | Acinetobacter sp. Tol 5 AtaA N-terminal half of C-terminal stalk fused to GCN4 adaptors (CstalkN) | | Descriptor: | Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WQA
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1ii) | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WOB
 
 | | Crystal structure of a prostate-specific WGA16 glycoprotein lectin, form I | | Descriptor: | hypothetical protein | | Authors: | Garenaux, E, Kanagawa, M, Tsuchiyama, T, Hori, K, Kanazawa, T, Goshima, A, Chiba, M, Yasue, H, Ikeda, A, Yamaguchi, Y, Sato, C, Kitajima, K. | | Deposit date: | 2013-12-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery, Primary, and Crystal Structures and Capacitation-related Properties of a Prostate-derived Heparin-binding Protein WGA16 from Boar Sperm
J.Biol.Chem., 290, 2015
|
|
1MPY
 
 | | STRUCTURE OF CATECHOL 2,3-DIOXYGENASE (METAPYROCATECHASE) FROM PSEUDOMONAS PUTIDA MT-2 | | Descriptor: | ACETONE, CATECHOL 2,3-DIOXYGENASE, FE (II) ION | | Authors: | Kita, A, Kita, S, Fujisawa, I, Inaka, K, Ishida, T, Horiike, K, Nozaki, M, Miki, K. | | Deposit date: | 1998-10-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Ppseudomonas putida mt-2.
Structure Fold.Des., 7, 1999
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
2H2N
 
 | | Crystal structure of human soluble calcium-activated nucleotidase (SCAN) with calcium ion | | Descriptor: | ACETATE ION, CALCIUM ION, Soluble calcium-activated nucleotidase 1 | | Authors: | Yang, M, Horii, K, Herr, A.B, Kirley, T.L. | | Deposit date: | 2006-05-19 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium-dependent dimerization of human soluble calcium activated nucleotidase: characterization of the dimer interface.
J.Biol.Chem., 281, 2006
|
|
2H2U
 
 | | Crystal structure of the E130Y mutant of human soluble calcium-activated nucleotidase (SCAN) with calcium ion | | Descriptor: | CALCIUM ION, Soluble calcium-activated nucleotidase 1 | | Authors: | Yang, M, Horii, K, Herr, A.B, Kirley, T.L. | | Deposit date: | 2006-05-19 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium-dependent dimerization of human soluble calcium activated nucleotidase: characterization of the dimer interface.
J.Biol.Chem., 281, 2006
|
|
1WMF
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WME
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.50 angstrom, 293 K) | | Descriptor: | CALCIUM ION, protease | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WMD
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.30 angstrom, 100 K) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WKY
 
 | | Crystal structure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Akita, M, Takeda, N, Hirasawa, K, Sakai, H, Kawamoto, M, Yamamoto, M, Grant, W.D, Hatada, Y, Ito, S, Horikoshi, K. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IC4
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1IC7
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1IC5
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
