2I0F
 
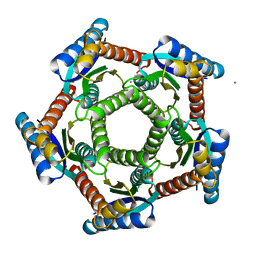 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2O6H
 
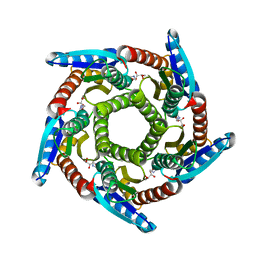 | | Lumazine synthase RibH1 from Brucella melitensis (Gene BMEI1187, Swiss-Prot entry Q8YGH2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2OBX
 
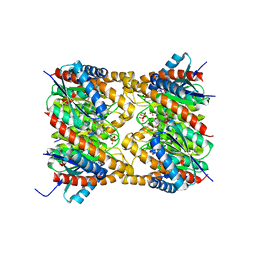 | | Lumazine synthase RibH2 from Mesorhizobium loti (Gene mll7281, Swiss-Prot entry Q986N2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-20 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and kinetic properties of lumazine synthase isoenzymes in the order rhizobiales
J.Mol.Biol., 373, 2007
|
|
3PQK
 
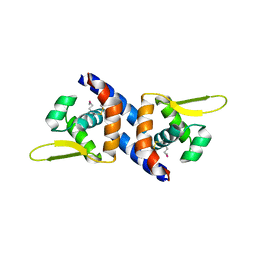 | |
3D5J
 
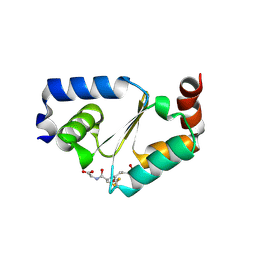 | | Structure of yeast Grx2-C30S mutant with glutathionyl mixed disulfide | | Descriptor: | GLUTATHIONE, Glutaredoxin-2, mitochondrial | | Authors: | Discola, K.F, de Oliveira, M.A, Barcena, J.A, Porras, P, Padilla, C.A, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae.
J.Mol.Biol., 385, 2009
|
|
2PO1
 
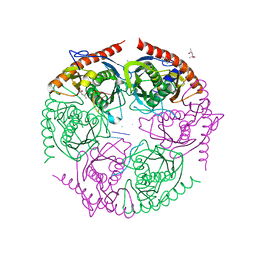 | |
2PO2
 
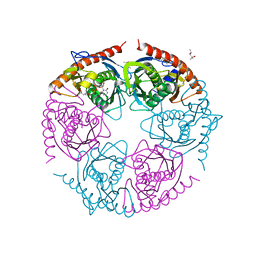 | |
2KDO
 
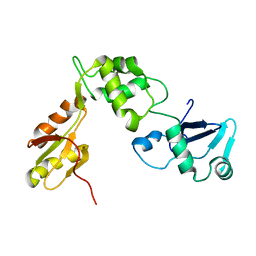 | | Structure of the human Shwachman-Bodian-Diamond syndrome protein, SBDS | | Descriptor: | Ribosome maturation protein SBDS | | Authors: | de Oliveira, J.F, Sforca, M.L, Blumenschein, T, Guimaraes, B.G, Zanchin, N.I.T, Zeri, A.C. | | Deposit date: | 2009-01-14 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and RNA interaction analysis of the human SBDS protein.
J.Mol.Biol., 396, 2010
|
|
2F59
 
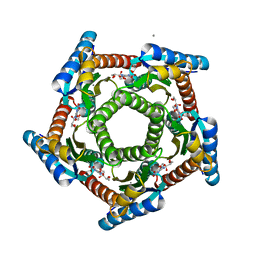 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
6OV6
 
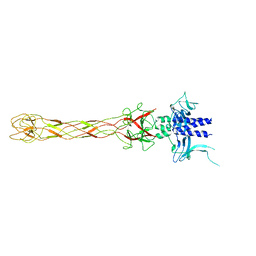 | | CRYSTALLOGRAPHIC STRUCTURE OF THE C24 PROTEIN FROM THE ANTARCTIC MICROORGANISM BIZIONIA ARGENTINENSIS | | Descriptor: | C24 PROTEIN, MANGANESE (II) ION | | Authors: | Klinke, S, Rinaldi, J, Guimaraes, B.G, Pellizza, L, Aran, M. | | Deposit date: | 2019-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the putative long tail fiber receptor-binding tip of a novel temperate bacteriophage from the Antarctic bacterium Bizionia argentinensis JUB59.
J.Struct.Biol., 212, 2020
|
|
2PO0
 
 | |
2PNZ
 
 | |
4RQI
 
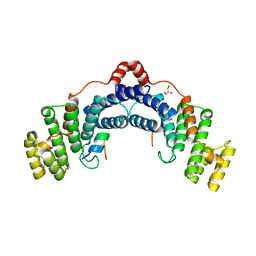 | | Structure of TRF2/RAP1 secondary interaction binding site | | Descriptor: | GLYCEROL, MAGNESIUM ION, Telomeric repeat-binding factor 2, ... | | Authors: | Miron, S, Guimaraes, B, Gaullier, G, Giraud-Panis, M.-J, Gilson, E, Le Du, M.-H. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4405 Å) | | Cite: | A higher-order entity formed by the flexible assembly of RAP1 with TRF2.
Nucleic Acids Res., 44, 2016
|
|
3D4M
 
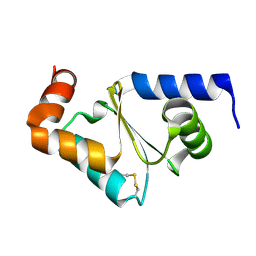 | | Glutaredoxin 2 oxidized structure | | Descriptor: | Glutaredoxin-2, mitochondrial | | Authors: | Discola, K.F, de Oliveira, M.A, Barcena, J.A, Porras, P, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2008-05-14 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae.
J.Mol.Biol., 385, 2009
|
|
3ER0
 
 | |
2REM
 
 | |
3H79
 
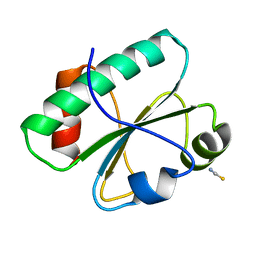 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
2F9L
 
 | | 3D structure of inactive human Rab11b GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAB11B, ... | | Authors: | Scapin, S.M.N, Guimaraes, B.G, Zanchin, N.I.T. | | Deposit date: | 2005-12-06 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform.
J.Struct.Biol., 154, 2006
|
|
2F9M
 
 | | 3D structure of active human Rab11b GTPase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Scapin, S.M.N, Guimaraes, B.G, Zanchin, N.I.T. | | Deposit date: | 2005-12-06 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform.
J.Struct.Biol., 154, 2006
|
|
2H7W
 
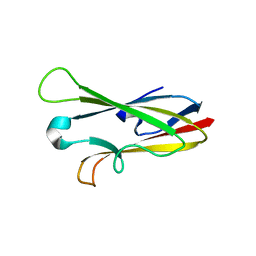 | |
3NRK
 
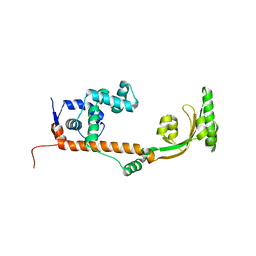 | | The crystal structure of the leptospiral hypothetical protein LIC12922 | | Descriptor: | LIC12922 | | Authors: | Giuseppe, P.O, Atzingen, M.V, Nascimento, A.L.T.O, Zanchin, N.I.T, Guimaraes, B.G. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of the leptospiral hypothetical protein LIC12922 reveals homology with the periplasmic chaperone SurA.
J.Struct.Biol., 173, 2011
|
|
4OWY
 
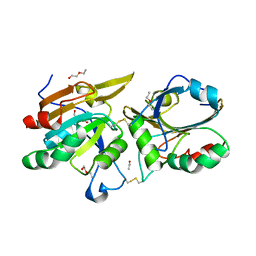 | | Crystal Structure of Ahp1 from Saccharomyces cerevisiae. Investigating the electron transfers. | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Peroxiredoxin type-2, ... | | Authors: | Schultz, L, Genu, V, Breyer, C.A, dos Santos, V.F, Guimaraes, B.G, Netto, L.E.S, de Oliveira, M.A. | | Deposit date: | 2014-02-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Ahp1 from Saccharomyces cerevisiae. Investigating the electron transfers.
To be published
|
|
4GQN
 
 | | Crystallographic structure of trimeric Riboflavin Synthase from Brucella abortus in complex with 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-08-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P3Z
 
 | | Chlamydia pneumoniae CopN (D29 construct) | | Descriptor: | CopN, GLYCEROL, SULFATE ION | | Authors: | Nawrotek, A, Guimaraes, B.G, Knossow, M, Gigant, B. | | Deposit date: | 2014-03-10 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae.
J.Biol.Chem., 289, 2014
|
|
4P40
 
 | | Chlamydia pneumoniae CopN | | Descriptor: | CHLORIDE ION, CopN, DIMETHYLAMINE, ... | | Authors: | Nawrotek, A, Guimaraes, B.G, Knossow, M, Gigant, B. | | Deposit date: | 2014-03-10 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae.
J.Biol.Chem., 289, 2014
|
|
