4KKS
 
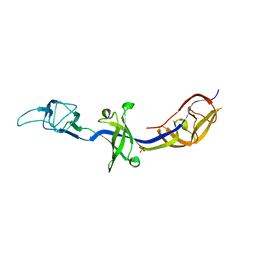 | | Crystal Structure of BesA (C2 form) | | Descriptor: | Membrane fusion protein, PHOSPHATE ION | | Authors: | Greene, N.P, Hinchliffe, P, Crow, A, Ababou, A, Hughes, C, Koronakis, V. | | Deposit date: | 2013-05-06 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an atypical periplasmic adaptor from a multidrug efflux pump of the spirochete Borrelia burgdorferi.
Febs Lett., 587, 2013
|
|
4KKU
 
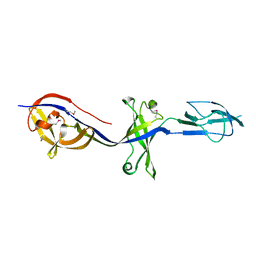 | | Structure of BesA (Selenomethinone derivative - P212121) | | Descriptor: | Membrane fusion protein | | Authors: | Greene, N.P, Hinchliffe, P, Crow, A, Ababou, A, Hughes, C, Koronakis, V. | | Deposit date: | 2013-05-06 | | Release date: | 2013-07-10 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of an atypical periplasmic adaptor from a multidrug efflux pump of the spirochete Borrelia burgdorferi.
Febs Lett., 587, 2013
|
|
4KKT
 
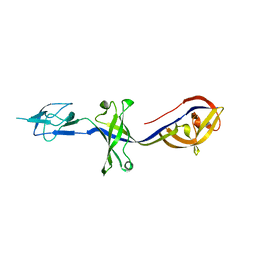 | | Crystal Structure of BesA (P21 form) | | Descriptor: | Membrane fusion protein | | Authors: | Greene, N.P, Hinchliffe, P, Crow, A, Ababou, A, Hughes, C, Koronakis, V. | | Deposit date: | 2013-05-06 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure of an atypical periplasmic adaptor from a multidrug efflux pump of the spirochete Borrelia burgdorferi.
Febs Lett., 587, 2013
|
|
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
2K8X
 
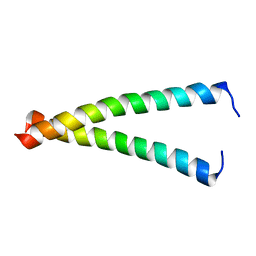 | |
1MV4
 
 | | TM9A251-284: A Peptide Model of the C-Terminus of a Rat Striated Alpha Tropomyosin | | Descriptor: | Tropomyosin 1 alpha chain | | Authors: | Greenfield, N.J, Swapna, G.V.T, Huang, Y, Palm, T, Graboski, S, Montelione, G.T, Hitchcock-Degregori, S.E. | | Deposit date: | 2002-09-24 | | Release date: | 2003-02-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Carboxyl Terminus of Striated alpha-Tropomyosin in Solution Reveals an Unusual Parallel Arrangement of Interacting alpha-Helices
Biochemistry, 42, 2003
|
|
1IHQ
 
 | | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B | | Descriptor: | CHIMERIC PEPTIDE GlyTM1bZip: TROPOMYOSIN ALPHA CHAIN, BRAIN-3 and GENERAL CONTROL PROTEIN GCN4 | | Authors: | Greenfield, N.J, Yuang, Y.J, Palm, T, Swapna, G.V, Monleon, D, Montelione, G.T, Hitchcock-Degregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein.
J.Mol.Biol., 312, 2001
|
|
1TMZ
 
 | | TMZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF ALPHA TROPOMYOSIN, NMR, 15 STRUCTURES | | Descriptor: | TMZIP | | Authors: | Greenfield, N.J, Montelione, G.T, Hitchcock-Degregori, S.E, Farid, R.S. | | Deposit date: | 1998-04-20 | | Release date: | 1998-06-17 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies.
Biochemistry, 37, 1998
|
|
1SLO
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLP
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, 16 STRUCTURES | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1997-04-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1LFU
 
 | | NMR Solution Structure of the Extended PBX Homeodomain Bound to DNA | | Descriptor: | 5'-D(*GP*CP*GP*CP*AP*TP*GP*AP*TP*TP*GP*CP*CP*C)-3', 5'-D(*GP*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*CP*GP*C)-3', homeobox protein PBX1 | | Authors: | Sprules, T, Green, N, Featherstone, M, Gehring, K. | | Deposit date: | 2002-04-12 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Lock and Key Binding of the HOX YPWM Peptide to the PBX Homeodomain
J.Biol.Chem., 278, 2003
|
|
5FTK
 
 | | Cryo-EM structure of human p97 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTL
 
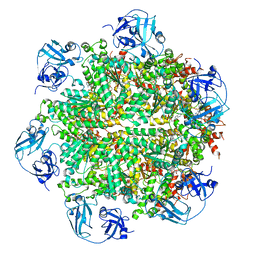 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTJ
 
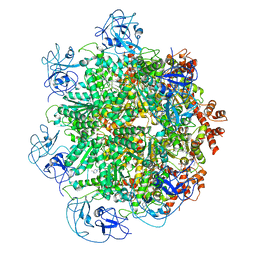 | | Cryo-EM structure of human p97 bound to UPCDC30245 inhibitor | | Descriptor: | 1-(3-(5-FLUORO-1H-INDOL-2-YL)PHENYL)PIPERIDIN-4-YL)(2-(4-ISOPROPYL-PIPERAZIN1-YL)ETHYL)-CARBAMATE, ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTN
 
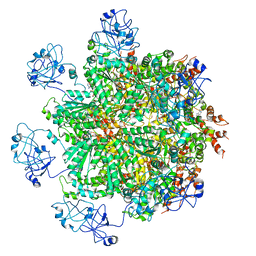 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation III) | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTM
 
 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
1MK5
 
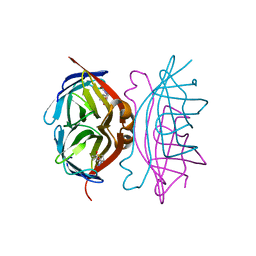 | | Wildtype Core-Streptavidin with Biotin at 1.4A. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Green, N.M, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
1MEP
 
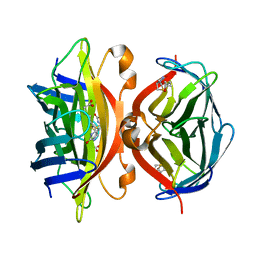 | | Crystal Structure of Streptavidin Double Mutant S45A/D128A with Biotin: Cooperative Hydrogen-Bond Interactions in the Streptavidin-Biotin System. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Stenkamp, R.E, Green, N.M, Stayton, P.S. | | Deposit date: | 2002-08-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
1HQT
 
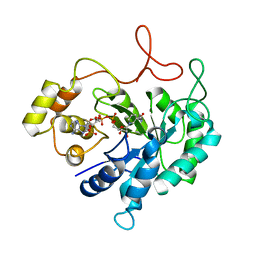 | | THE CRYSTAL STRUCTURE OF AN ALDEHYDE REDUCTASE Y50F MUTANT-NADP COMPLEX AND ITS IMPLICATIONS FOR SUBSTRATE BINDING | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Hyndman, D, Green, N.C, Li, L, Korithoski, B, Jia, Z, Flynn, T.G. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of an Aldehyde Reductase Y50F Mutant-NADP Complex and its Implications for Substrate Binding
Chem.Biol.Interact., 132, 2001
|
|
1DU6
 
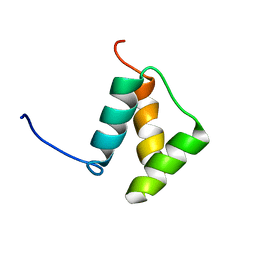 | |
4WHN
 
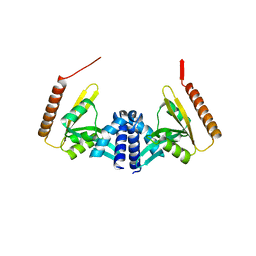 | | Structure of toxin-activating acyltransferase (TAAT) | | Descriptor: | ApxC, CITRIC ACID | | Authors: | Crow, A, Greene, N.P, Hughes, C, Koronakis, V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a bacterial toxin-activating acyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7Z6W
 
 | | Complex of E. coli LolA and lipoprotein | | Descriptor: | Outer-membrane lipoprotein carrier protein, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Kaplan, E, Greene, N.P, Koronakis, V. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis of lipoprotein recognition by the bacterial Lol trafficking chaperone LolA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z6X
 
 | | Complex of E. coli LolA R43L mutant and lipoprotein | | Descriptor: | Outer-membrane lipoprotein carrier protein, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Kaplan, E, Greene, N.P, Koronakis, V. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of lipoprotein recognition by the bacterial Lol trafficking chaperone LolA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6FHM
 
 | | Crystal structure of the F47E mutant of the lipoprotein localization factor, LolA | | Descriptor: | GLYCEROL, Outer-membrane lipoprotein carrier protein | | Authors: | Kaplan, E, Greene, N.P, Crow, A, Koronakis, V. | | Deposit date: | 2018-01-15 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4TKO
 
 | | Structure of the periplasmic adaptor protein EmrA | | Descriptor: | EmrA, ISOPROPYL ALCOHOL, MAGNESIUM ION | | Authors: | Hinchliffe, P, Greene, N.P, Paterson, N.G, Crow, A, Hughes, C, Koronakis, V. | | Deposit date: | 2014-05-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump.
Febs Lett., 588, 2014
|
|
