1TC1
 
 | | A 1.4 ANGSTROM CRYSTAL STRUCTURE FOR THE HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE OF TRYPANOSOMA CRUZI | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMYCIN B, PROTEIN (HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE) | | Authors: | Focia, P.J, Craig III, S.P, Nieves-Alicea, R, Fletterick, R.J, Eakin, A.E. | | Deposit date: | 1998-09-30 | | Release date: | 1999-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
Biochemistry, 37, 1998
|
|
1TC2
 
 | | TERNARY SUBSTRATE COMPLEX OF THE HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE FROM TRYPANOSOMA CRUZI | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, MAGNESIUM ION, ... | | Authors: | Focia, P.J, Craig III, S.P, Eakin, A.E. | | Deposit date: | 1998-11-04 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Approaching the transition state in the crystal structure of a phosphoribosyltransferase.
Biochemistry, 37, 1998
|
|
1OKK
 
 | |
2CNW
 
 | | GDPALF4 complex of the SRP GTPases Ffh and FtsY | | Descriptor: | CELL DIVISION PROTEIN FTSY, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Focia, P.J, Gawronski-Salerno, J, Coon V, J.S, Freymann, D.M. | | Deposit date: | 2006-05-24 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a Gdp:Alf(4) Complex of the Srp Gtpases Ffh and Ftsy, and Identification of a Peripheral Nucleotide Interaction Site.
J.Mol.Biol., 360, 2006
|
|
2IYL
 
 | | Structure of an FtsY:GDP complex | | Descriptor: | CELL DIVISION PROTEIN FTSY, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Focia, P.J, Freymann, D.M. | | Deposit date: | 2006-07-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of the T. aquaticus FtsY:GDP complex suggests functional roles for the C-terminal helix of the SRP GTPases.
Proteins, 66, 2007
|
|
4UUJ
 
 | | POTASSIUM CHANNEL KCSA-FAB WITH TETRAHEXYLAMMONIUM | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Burdette, D, Wagner, T, Focia, P.J, Gross, A. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Kcsa in Complex with Symmetrical Quaternary Ammonium Compounds Reveal a Hydrophobic Binding Site.
Biochemistry, 53, 2014
|
|
6D71
 
 | | Crystal Structure of the Human Miro1 N-terminal GTPase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 1 | | Authors: | Smith, K.P, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2018-04-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7180779 Å) | | Cite: | Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase.
J.Struct.Biol., 212, 2020
|
|
1MXO
 
 | | AmpC beta-lactamase in complex with an m.carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Morandi, F, Caselli, E, Morandi, S, Focia, P.J, Blazquez, J, Shoichet, B.K, Prati, F. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Nanomolar inhibitors of AmpC beta-lactamase.
J.Am.Chem.Soc., 125, 2003
|
|
1MY8
 
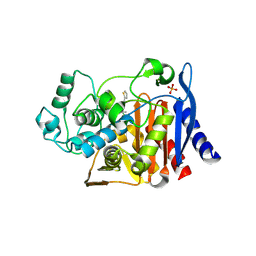 | | AmpC beta-lactamase in complex with an M.carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, PHOSPHATE ION, beta-lactamase | | Authors: | Morandi, F, Caselli, E, Morandi, S, Focia, P.J, Blazquez, J, Shoichet, B.K, Prati, F. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Nanomolar inhibitors of AmpC beta-lactamase.
J.Am.Chem.Soc., 125, 2003
|
|
3B3Q
 
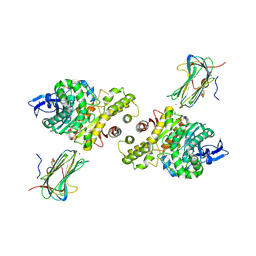 | | Crystal structure of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, X, Liu, H, Shim, A, Focia, P, He, X. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3IFX
 
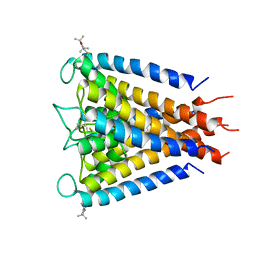 | | Crystal structure of the Spin-labeled KcsA mutant V48R1 | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, TETRABUTYLAMMONIUM ION, ... | | Authors: | Cieslak, J.A, Focia, P.J, Gross, A. | | Deposit date: | 2009-07-26 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | EPR (3.56 Å), X-RAY DIFFRACTION | | Cite: | Electron Spin-Echo Envelope Modulation (ESEEM) Reveals Water and Phosphate Interactions with the KcsA Potassium Channel
Biochemistry, 49, 2010
|
|
2O27
 
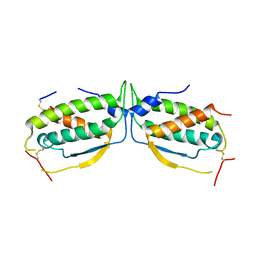 | |
3EJJ
 
 | | Structure of M-CSF bound to the first three domains of FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Colony stimulating factor-1, Macrophage colony-stimulating factor 1 receptor | | Authors: | Chen, X, Liu, H, Focia, P.J, Shim, A, He, X. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VB2
 
 | | Crystal structure of Cu(I)CusF | | Descriptor: | CATION EFFLUX SYSTEM PROTEIN CUSF, COPPER (II) ION, SULFATE ION | | Authors: | Xue, Y, Davis, A.V, Balakrishnan, G, Stasser, J.P, Staehlin, B.M, Focia, P, Spiro, T.G, Penner-Hahn, J.E, O'Halloran, T.V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cu(I) Recognition Via Cation-Pi and Methionine Interactions in Cusf.
Nat.Chem.Biol., 4, 2008
|
|
2VB3
 
 | | Crystal structure of Ag(I)CusF | | Descriptor: | CATION EFFLUX SYSTEM PROTEIN CUSF, SILVER ION | | Authors: | Xue, Y, Davis, A.V, Balakrishnan, G, Stasser, J.P, Staehlin, B.M, Focia, P, Spiro, T.G, Penner-Hahn, J.E, O'Halloran, T.V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Cu(I) Recognition Via Cation-Pi and Methionine Interactions in Cusf.
Nat.Chem.Biol., 4, 2008
|
|
6UHN
 
 | | Crystal Structure of C148 mGFP-cDNA-1 | | Descriptor: | C148 mGFP-cDNA-1, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHR
 
 | | Crystal Structure of C148 mGFP-scDNA-2 | | Descriptor: | C148 mGFP-scDNA-2 | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
2O26
 
 | | Structure of a class III RTK signaling assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Kit ligand, Mast/stem cell growth factor receptor, ... | | Authors: | Liu, H, Chen, X, Focia, P.J, He, X. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases.
Embo J., 26, 2007
|
|
6UHQ
 
 | | Crystal Structure of C148 mGFP-cDNA-3 | | Descriptor: | C148 mGFP-cDNA-3, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHK
 
 | | Crystal Structure of C176 mGFP | | Descriptor: | C176 mGFP | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHJ
 
 | | X-ray Structure of C148 mGFP | | Descriptor: | C148 mGFP | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHP
 
 | | Crystal Structure of C148 mGFP-ncDNA-1 | | Descriptor: | C148 mGFP-ncDNA-1 | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHL
 
 | | Crystal Structure of C148 mGFP-scDNA-1 | | Descriptor: | C148 mGFP-scDNA-1, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHM
 
 | | Crystal Structure of a Physical Mixture of C148 mGFP and scDNA-1 | | Descriptor: | C148 mGFP | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHO
 
 | | Crystal Structure of C148 mGFP-cDNA-2 | | Descriptor: | C148 mGFP-cDNA-2, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
