2VC5
 
 | | Structural basis for natural lactonase and promiscuous phosphotriesterase activities | | Descriptor: | 1,2-ETHANEDIOL, ARYLDIALKYLPHOSPHATASE, COBALT (II) ION, ... | | Authors: | Elias, M, Dupuy, J, Merone, L, Mandrich, L, Moniot, S, Lecomte, C, Rossi, M, Masson, P, Manco, G, Chabriere, E. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities.
J.Mol.Biol., 379, 2008
|
|
2VC7
 
 | | Structural basis for natural lactonase and promiscuous phosphotriesterase activities | | Descriptor: | (4S)-4-(decanoylamino)-5-hydroxy-3,4-dihydro-2H-thiophenium, 1,2-ETHANEDIOL, ARYLDIALKYLPHOSPHATASE, ... | | Authors: | Elias, M, Dupuy, J, Merone, L, Mandrich, L, Moniot, S, Rochu, D, Lecomte, C, Rossi, M, Masson, P, Manco, G, Chabriere, E. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities.
J.Mol.Biol., 379, 2008
|
|
3UF9
 
 | | Crystal structure of SsoPox in complex with the phosphotriester fensulfothion | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (II) ION, ... | | Authors: | Elias, M, Gotthard, G, Hiblot, J, Chabriere, E. | | Deposit date: | 2011-10-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Characterisation of the organophosphate hydrolase catalytic activity of SsoPox
Sci Rep, 2, 2012
|
|
4F18
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 8.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
4F19
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 4.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
6XIX
 
 | |
6XJE
 
 | | Triuret Hydrolase (TrtA) from Herbaspirillum sp. BH-1 C162S bound with triuret | | Descriptor: | 1,2-ETHANEDIOL, Cysteine hydrolase, tricarbonodiimidic diamide | | Authors: | Tassoulas, L.T, Elias, M.H, Wackett, L.P. | | Deposit date: | 2020-06-23 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of an ultraspecific triuret hydrolase (TrtA) establishes the triuret biodegradation pathway.
J.Biol.Chem., 296, 2020
|
|
6CGY
 
 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to a phosphate anion | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
6CGZ
 
 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to C6-AHL | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
6CH0
 
 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to a glycerol molecule | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
6XJ4
 
 | | Triuret Hydrolase (TrtA) from Herbaspirillum sp. BH-1 C162S bound with biuret | | Descriptor: | 1,2-ETHANEDIOL, Cysteine hydrolase, dicarbonimidic diamide | | Authors: | Tassoulas, L.T, Elias, M.H, Wackett, L.P. | | Deposit date: | 2020-06-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of an ultraspecific triuret hydrolase (TrtA) establishes the triuret biodegradation pathway.
J.Biol.Chem., 296, 2020
|
|
4Q8R
 
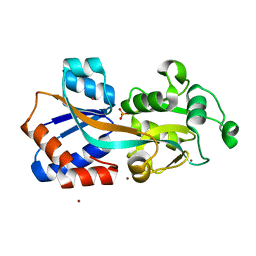 | | Crystal structure of a Phosphate Binding Protein (PBP-1) from Clostridium perfringens | | Descriptor: | PHOSPHATE ION, Phosphate ABC transporter, phosphate-binding protein, ... | | Authors: | Gonzalez, D, Richez, M, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2014-04-28 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the phosphate-binding protein (PBP-1) of an ABC-type phosphate transporter from Clostridium perfringens.
Sci Rep, 4, 2014
|
|
2V3Q
 
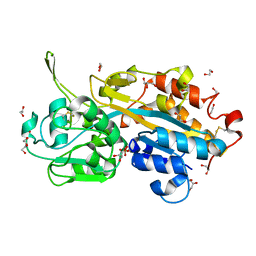 | | Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, HUMAN PHOSPHATE BINDING PROTEIN, ... | | Authors: | Morales, R, Berna, A, Carpentier, P, Elias, M, Contreras-Martel, C, Renault, F, Nicodeme, M, Chesne-Seck, M.-L, Bernier, F, Dupuy, J, Schaeffer, C, Diemer, H, Van Dorsselaer, A, Fontecilla, J.C, Masson, P, Rochu, D, Chabriere, E. | | Deposit date: | 2007-06-20 | | Release date: | 2008-07-22 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
6XK1
 
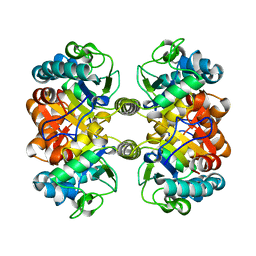 | |
6XJM
 
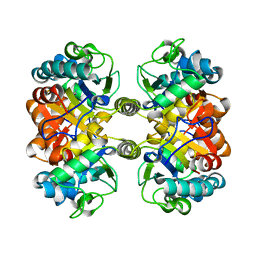 | |
4RDY
 
 | | Crystal structure of VmoLac bound to 3-oxo-C10 AHL | | Descriptor: | 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide, COBALT (II) ION, GLYCEROL, ... | | Authors: | Hiblot, J, Bzdrenga, J, Champion, C, Gotthard, G, Gonzalez, D, Chabriere, E, Elias, M. | | Deposit date: | 2014-09-20 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia.
Sci Rep, 5, 2015
|
|
7LTE
 
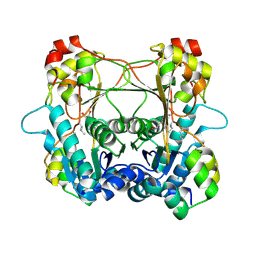 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA) heteromeric complex (with SAH) | | Descriptor: | LigA domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTH
 
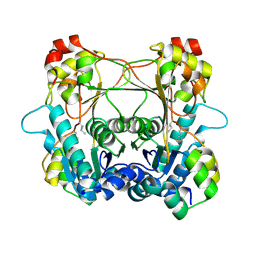 | | Structure of the alpha-N-methyltransferase (SonM mutant Y93F) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTS
 
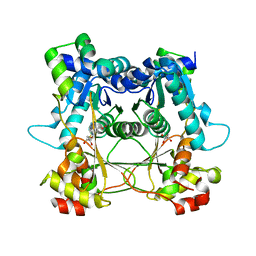 | | Structure of the alpha-N-methyltransferase (SonM mutant R67A) and RiPP precursor (SonA) heteromeric complex (with SAH) | | Descriptor: | LigA domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTR
 
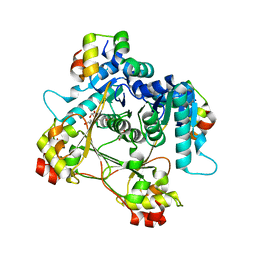 | | Structure of the heteromeric complex between the alpha-N-methyltransferase (SonM) and a truncated construct of the RiPP precursor (SonA) (with SAM) | | Descriptor: | GLYCEROL, LigA domain-containing protein, S-ADENOSYLMETHIONINE, ... | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTC
 
 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTF
 
 | | Structure of the alpha-N-methyltransferase (SonM mutant Y58F) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
8T1S
 
 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA with QSY deletion) heteromeric complex (bound to SAH) | | Descriptor: | Alpha-N-methyltransferase, Extradiol ring-cleavage dioxygenase LigAB LigA subunit domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Crone, K.K, Jomori, T, Miller, F.S, Gralnick, J, Elias, M, Freeman, M.F. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RiPP enzyme heterocomplex structure-guided discovery of a bacterial borosin alpha- N -methylated peptide natural product.
Rsc Chem Biol, 4, 2023
|
|
8T1T
 
 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA with QSY deletion) heteromeric complex (bound to SAM) | | Descriptor: | Alpha-N-methyltransferase (SonM), RiPP precursor (SonA), S-ADENOSYLMETHIONINE, ... | | Authors: | Crone, K.K, Jomori, T, Miller, F.S, Gralnick, J, Elias, M, Freeman, M.F. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | RiPP enzyme heterocomplex structure-guided discovery of a bacterial borosin alpha- N -methylated peptide natural product.
Rsc Chem Biol, 4, 2023
|
|
4RE0
 
 | | Crystal structure of VmoLac in P622 space group | | Descriptor: | COBALT (II) ION, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Hiblot, J, Bzdrenga, J, Champion, C, Gotthard, G, Gonzalez, D, Chabriere, E, Elias, M. | | Deposit date: | 2014-09-20 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia.
Sci Rep, 5, 2015
|
|
