5D6V
 
 | |
4RBU
 
 | |
4RBT
 
 | |
4RBV
 
 | |
1M3H
 
 | | Crystal Structure of Hogg1 D268E Mutant with Product Oligonucleotide | | Descriptor: | 5'-D(P*GP*CP*GP*TP*CP*CP*AP*(DDX))-3', 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*TP*AP*CP*C)-3', ... | | Authors: | Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-06-27 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of End Products Resulting from Lesion Processing by a DNA Glycosylase/Lyase
Chem.Biol., 11, 2004
|
|
7WRS
 
 | |
7WRU
 
 | |
4DCP
 
 | | Crystal Structure of caspase 3, L168F mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4DCJ
 
 | | Crystal structure of caspase 3, L168D mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4DCO
 
 | | Crystal Structure of caspase 3, L168Y mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
1M3Q
 
 | |
1MQ0
 
 | | Crystal Structure of Human Cytidine Deaminase | | Descriptor: | 1-BETA-RIBOFURANOSYL-1,3-DIAZEPINONE, Cytidine Deaminase, ZINC ION | | Authors: | Chung, S.J, Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-09-13 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human cytidine deaminase bound to a potent inhibitor
J.Med.Chem., 48, 2005
|
|
2K0G
 
 | |
2KXL
 
 | |
7EV9
 
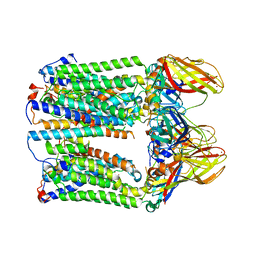 | | cryoEM structure of particulate methane monooxygenase associated with Cu(I) | | Descriptor: | Ammonia monooxygenase/methane monooxygenase, subunit C family protein, COPPER (I) ION, ... | | Authors: | Chang, W.H, Lin, H.H, Tsai, I.K, Huang, S.H, Chung, S.C, Tu, I.P, Yu, S.F, Chan, S.I. | | Deposit date: | 2021-05-20 | | Release date: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Copper Centers in the Cryo-EM Structure of Particulate Methane Monooxygenase Reveal the Catalytic Machinery of Methane Oxidation.
J.Am.Chem.Soc., 143, 2021
|
|
6LDK
 
 | |
3B97
 
 | | Crystal Structure of human Enolase 1 | | Descriptor: | Alpha-enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Kang, H.J, Jung, S.K, Kim, S.J, Chung, S.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2H80
 
 | | NMR structures of SAM domain of Deleted in Liver Cancer 2 (DLC2) | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Li, H.Y, Fung, K.L, Jin, D.Y, Chung, S.S, Ko, B.C, Sun, H.Z. | | Deposit date: | 2006-06-06 | | Release date: | 2007-05-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2
Proteins, 67, 2007
|
|
3Q9U
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | COENZYME A, CoA binding protein, consensus ankyrin repeat | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3FCI
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{(E)-[(3-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}propoxy)imino]methyl}benzoic acid, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCF
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-[(1E,7E)-8-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-3,6-dioxa-2,7-diazaocta-1,7-dien-1-yl]benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3FCL
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{[(4-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}butyl)amino]methyl}benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
1HDU
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HEE
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with L-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, L-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
