4WCM
 
 | |
4WCN
 
 | | Crystal Structure of Tripeptide bound Cell Shape Determinant Csd4 protein from Helicobacter pylori | | Descriptor: | Conserved hypothetical secreted protein, IODIDE ION, N-acetyl-L-alanyl-N-[(1S,5R)-5-amino-1,5-dicarboxypentyl]-D-glutamine, ... | | Authors: | Chan, A.C, Murphy, M.E. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Helical Shape of Helicobacter pylori Requires an Atypical Glutamine as a Zinc Ligand in the Carboxypeptidase Csd4.
J.Biol.Chem., 290, 2015
|
|
4WCK
 
 | |
4WCL
 
 | |
8T4C
 
 | |
8D27
 
 | |
6P5T
 
 | | Surface-layer (S-layer) RsaA protein from Caulobacter crescentus bound to strontium and iodide | | Descriptor: | IODIDE ION, S-layer protein, STRONTIUM ION | | Authors: | Chan, A.C, Herrmann, J, Smit, J, Wakatsuki, S, Murphy, M.E. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bacterial surface layer protein exploits multistep crystallization for rapid self-assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8G64
 
 | | Heme-bound flavodoxin FldH from Fusobacterium nucleatum | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chan, A.C, Wolthers, K.R, Murphy, M.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new member of the flavodoxin superfamily from Fusobacterium nucleatum that functions in heme trafficking and reduction of anaerobilin.
J.Biol.Chem., 299, 2023
|
|
2G5G
 
 | |
6XI7
 
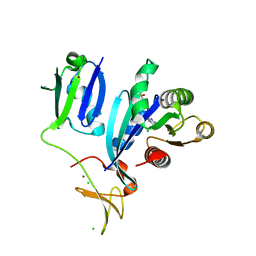 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form I) | | Descriptor: | CHLORIDE ION, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Chan, A.H, Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
8CZA
 
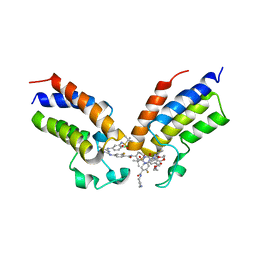 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to GXH-IV-075 | | Descriptor: | 4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluoro-N-[1-(14-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]phenyl}-14-oxo-4,7,10-trioxa-13-azatetradecanan-1-oyl)piperidin-4-yl]benzamide, Bromodomain testis-specific protein, SODIUM ION | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
8EBZ
 
 | |
6PPA
 
 | | Crystal structure of the unliganded bromodomain of human BRD7 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, PHOSPHATE ION | | Authors: | Chan, A, Karim, M.R, Zhu, J, Schonbrunn, E. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
7BJY
 
 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to Ro3280 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Isoform 2 of Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LRO
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LP0
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7MR5
 
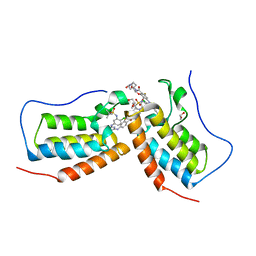 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-052 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-{oxybis[(ethane-2,1-diyl)oxy(1-oxoethane-2,1-diyl)piperidine-1,4-diyl]}bis{4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluorobenzamide} | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
5I0V
 
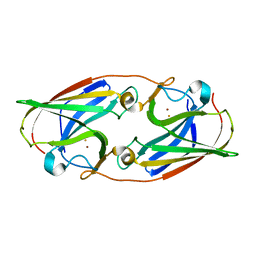 | |
5I0W
 
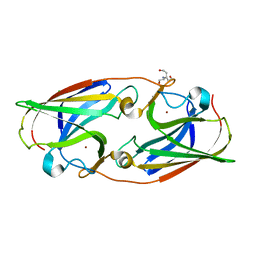 | |
7MR8
 
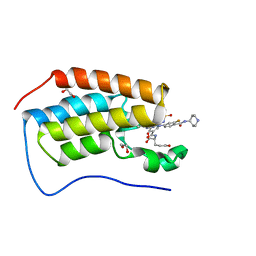 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-076 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]benzoyl}-L-glutamic acid | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-076
To Be Published
|
|
7MRC
 
 | |
5I0X
 
 | |
5I0Y
 
 | |
6WEF
 
 | |
