2M46
 
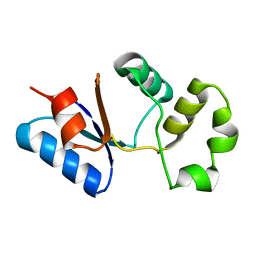 | | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841 | | Descriptor: | Arsenate reductase, putative | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841
To be Published
|
|
7K1O
 
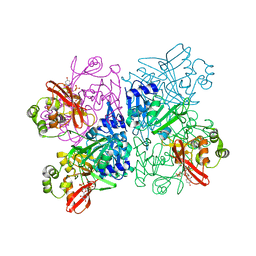 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1-(3,5-di-O-phosphono-alpha-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione, Uridylate-specific endoribonuclease | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Welk, L, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-08 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate
To Be Published
|
|
6VWW
 
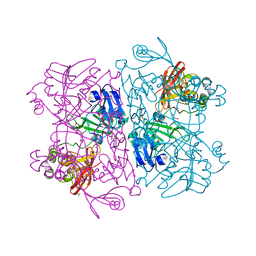 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6P1J
 
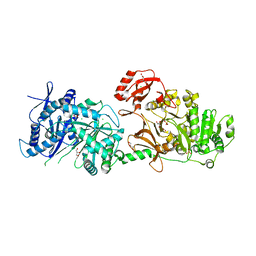 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo2 serine module | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-20 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
5DVY
 
 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
6P3I
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with Mg | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-05 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
7M1Y
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with ebselen | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Maltseva, N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-03-15 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with ebselen
to be published
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6QKY
 
 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6U69
 
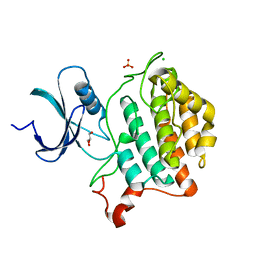 | | Crystal structure of Yck2 from Candida albicans, apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Overcoming Fungal Echinocandin Resistance through Inhibition of the Non-essential Stress Kinase Yck2.
Cell Chem Biol, 27, 2020
|
|
7SWX
 
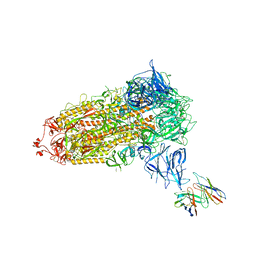 | |
6MGU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate
To Be Published
|
|
5IBX
 
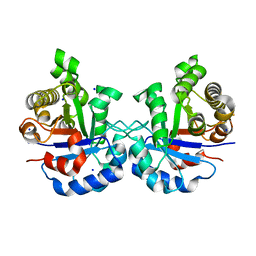 | | 1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae
To Be Published
|
|
7JFQ
 
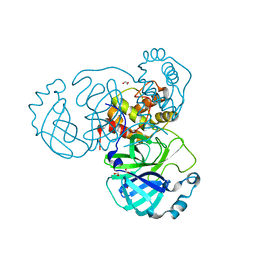 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
5W6L
 
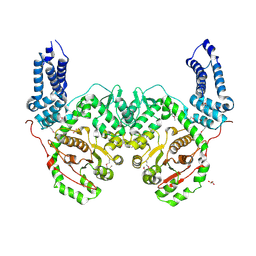 | | Crystal Structure of RRSP, a MARTX Toxin Effector Domain from Vibrio vulnificus CMCP6 | | Descriptor: | CHLORIDE ION, GLYCEROL, RTX repeat-containing cytotoxin, ... | | Authors: | Minasov, G, Wawrzak, Z, Biancucci, M, Shuvalova, L, Dubrovska, I, Satchell, K.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The bacterial Ras/Rap1 site-specific endopeptidase RRSP cleaves Ras through an atypical mechanism to disrupt Ras-ERK signaling.
Sci Signal, 11, 2018
|
|
6OYF
 
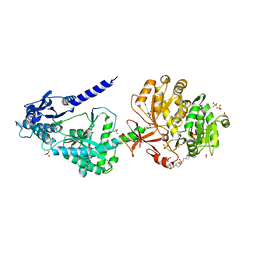 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
5EAV
 
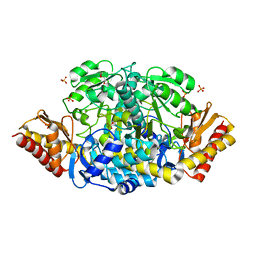 | | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii
To Be Published
|
|
5E5I
 
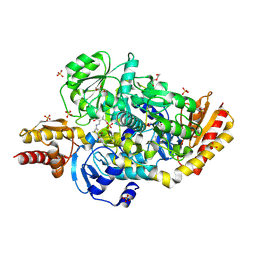 | | Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator | | Descriptor: | 4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-4-enoic acid, 6-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-4-oxidanylidene-hexanoic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator.
To Be Published
|
|
5E3K
 
 | | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid | | Descriptor: | 4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-4-enoic acid, Aminotransferase, CARBONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-02 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid
To Be Published
|
|
6OX6
 
 | | Crystal structure of the complex between the Type VI effector Tas1 and its immunity protein | | Descriptor: | ACETATE ION, PA14_01140, Tas1 | | Authors: | Ahmad, S, Stogios, P.J, Skarina, T, Whitney, J, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-13 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp.
Nature, 575, 2019
|
|
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
5WI5
 
 | | 2.0 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MAGNESIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-07-18 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
5JPH
 
 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus in complex with CoA | | Descriptor: | Acetyltransferase SACOL1063, CHLORIDE ION, COENZYME A | | Authors: | Majorek, K.A, Osinski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-06-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
