1CHC
 
 | |
1HFH
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH AND 16TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1HFI
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1VVC
 
 | |
1VVD
 
 | |
3OXU
 
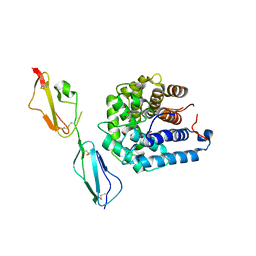 | | Complement components factor H CCP19-20 and C3d in complex | | Descriptor: | Complement C3, GLYCEROL, HF protein | | Authors: | Morgan, H.P, Schmidt, C.Q, Guariento, M, Gillespie, D, Herbert, A.P, Mertens, H, Blaum, B.S, Svergun, D, Johansson, C.M, Uhrin, D, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2010-09-22 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for engagement by complement factor H of C3b on a self surface.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1HCC
 
 | |
4B2S
 
 | | Solution structure of CCP modules 11-12 of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Makou, E, Mertens, H.D, Maciejewski, M, Soares, D.C, Matis, I, Schmidt, C.Q, Herbert, A.P, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ccp Modules 10-12 Illuminates Functional Architecture of the Complement Regulator, Factor H.
J.Mol.Biol., 424, 2012
|
|
1GKG
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-14 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
2WCY
 
 | | NMR solution structure of factor I-like modules of complement C7. | | Descriptor: | COMPLEMENT COMPONENT C7 | | Authors: | Phelan, M.M, Thai, C.T, Soares, D.C, Ogata, R.T, Barlow, P.N, Bramham, J. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Factor I-Like Modules from Complement C7 Reveals a Pair of Follistatin Domains in Compact Pseudosymmetric Arrangement.
J.Biol.Chem., 284, 2009
|
|
2JGX
 
 | | Structure of CCP module 7 of complement factor H - The AMD Not at risk varient (402Y) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
2JGW
 
 | | Structure of CCP module 7 of complement factor H - The AMD at risk varient (402H) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
1DV9
 
 | | STRUCTURAL CHANGES ACCOMPANYING PH-INDUCED DISSOCIATION OF THE B-LACTOGLOBULIN DIMER | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Uhrinova, S, Smith, M.H, Jameson, G.B, Uhrin, D, Sawyer, L, Barlow, P.N. | | Deposit date: | 2000-01-20 | | Release date: | 2000-02-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural changes accompanying pH-induced dissociation of the beta-lactoglobulin dimer.
Biochemistry, 39, 2000
|
|
3SW0
 
 | | Structure of the C-terminal region (modules 18-20) of complement regulator Factor H | | Descriptor: | Complement factor H, GLYCEROL, PHOSPHATE ION | | Authors: | Morgan, H.P, Guariento, M, Schmidt, C.Q, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of the C-Terminal Region (Modules 18-20) of Complement Regulator Factor H (FH).
Plos One, 7, 2012
|
|
3R62
 
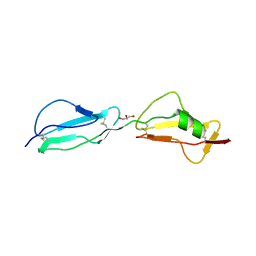 | | Structure of complement regulator Factor H mutant, T1184R. | | Descriptor: | Complement factor H, GLYCEROL | | Authors: | Morgan, H.P, Jiang, J, Herbert, A.P, Kavanagh, D, Uhran, D, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic determination of the disease-associated T1184R variant of the complement Factor H.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2MCZ
 
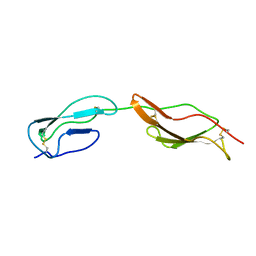 | | CR1 Sushi domains 1 and 2 | | Descriptor: | Complement receptor type 1 | | Authors: | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
1Z1M
 
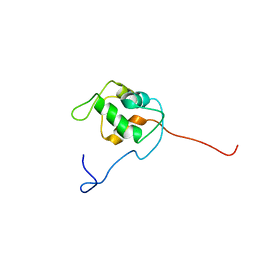 | | NMR structure of unliganded MDM2 | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Uhrinova, S, Uhrin, D, Powers, H, Watt, K, Zheleva, D, Fischer, P, McInnes, C, Barlow, P.N. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Free MDM2 N-terminal Domain Reveals Conformational Adjustments that Accompany p53-binding
J.Mol.Biol., 350, 2005
|
|
2MCY
 
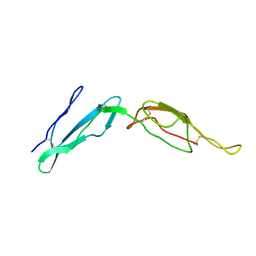 | | CR1 Sushi domains 2 and 3 | | Descriptor: | Complement receptor type 1 | | Authors: | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2014-01-22 | | Method: | SOLUTION NMR | | Cite: | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
2RLP
 
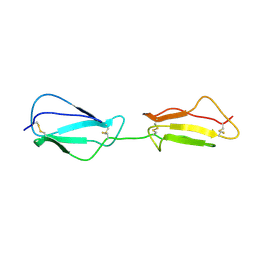 | | NMR structure of CCP modules 1-2 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-28 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
2V8E
 
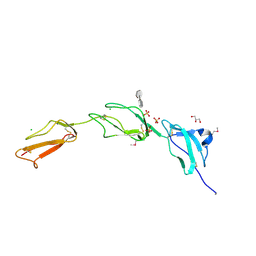 | | Crystal structure of Human Complement Factor H, SCR domains 6-8 (H402 risk variant), in complex with ligand. | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CHLORIDE ION, COMPLEMENT FACTOR H, ... | | Authors: | Prosser, B.E, Johnson, S, Roversi, P, Herbert, A.P, Blaum, B.S, Tyrrell, J, Jowitt, T.A, Clark, S.J, Tarelli, E, Uhrin, D, Barlow, P.N, Sim, R.B, Day, A.J, Lea, S.M. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Complement Factor H Linked Age-Related Macular Degeneration.
J.Exp.Med., 204, 2007
|
|
2UWN
 
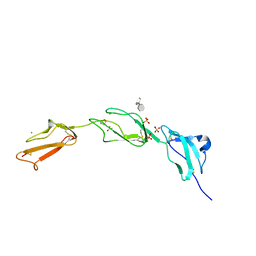 | | Crystal structure of Human Complement Factor H, SCR domains 6-8 (H402 risk variant), in complex with ligand. | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Prosser, B.E, Johnson, S, Roversi, P, Herbert, A.P, Blaum, B.S, Tyrrell, J, Jowitt, T.A, Clark, S.J, Terelli, E, Uhrin, D, Barlow, P.N, Sim, R.B, Day, A.J, Lea, S.M. | | Deposit date: | 2007-03-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Complement Factor H Linked Age-Related Macular Degeneration.
J.Exp.Med., 204, 2007
|
|
2KMS
 
 | | Combined high- and low-resolution techniques reveal compact structure in central portion of factor H despite long inter-modular linkers | | Descriptor: | Complement factor H | | Authors: | Schmidt, C.Q, Herbert, A.P, Guariento, M, Mertens, H.D.T, Soares, D.C, Uhrin, D, Rowe, A.J, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2009-08-04 | | Release date: | 2009-11-03 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | The Central Portion of Factor H (Modules 10-15) Is Compact and Contains a Structurally Deviant CCP Module
J.Mol.Biol., 395, 2010
|
|
2RLQ
 
 | | NMR structure of CCP modules 2-3 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-29 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
1GKN
 
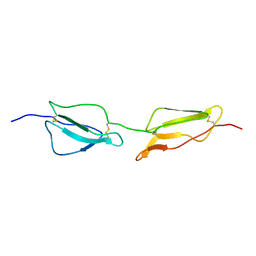 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
2BZM
 
 | | Solution structure of the primary host recognition region of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Uhrin, D, Lyon, M, Pangburn, M.K, Barlow, P.N. | | Deposit date: | 2005-08-18 | | Release date: | 2006-03-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Disease-Associated Sequence Variations Congregate in a Polyanion Recognition Patch on Human Factor H Revealed in Three-Dimensional Structure.
J.Biol.Chem., 281, 2006
|
|
