1F60
 
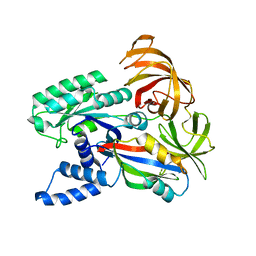 | | CRYSTAL STRUCTURE OF THE YEAST ELONGATION FACTOR COMPLEX EEF1A:EEF1BA | | Descriptor: | ELONGATION FACTOR EEF1A, ELONGATION FACTOR EEF1BA | | Authors: | Andersen, G.R, Pedersen, L, Valente, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2000-06-19 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha.
Mol.Cell, 6, 2000
|
|
9EQ4
 
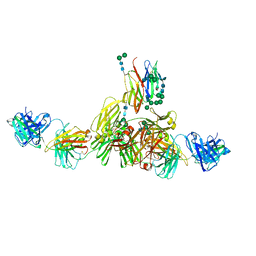 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5
To be published
|
|
9EQ3
 
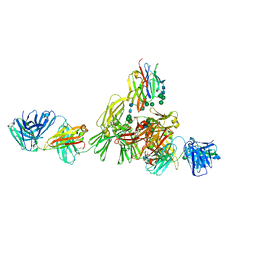 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8
To be published
|
|
8CEM
 
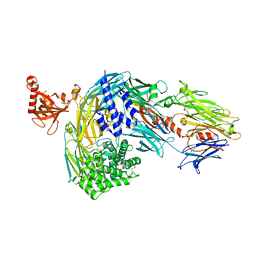 | | Structure of bovine native C3, re-refinement | | Descriptor: | Complement C3 alpha chain, Complement C3 beta chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Andersen, G.R, Fredslund, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of bovine complement component 3 reveals the basis for thioester function.
J Mol Biol, 361, 2006
|
|
8C1B
 
 | |
8C1C
 
 | | Structure of IgE bound to the ectodomain of FceRIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of IgE bound to the ectodomain of FceRIa
To Be Published
|
|
8COE
 
 | | complement C5 in complex with the LCP0195 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5 alpha chain, Complement C5 beta chain, ... | | Authors: | Andersen, G.R, Pedersen, D.V. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Characterization of the bispecific VHH antibody gefurulimab (ALXN1720) targeting complement component 5, and designed for low volume subcutaneous administration.
Mol.Immunol., 165, 2023
|
|
1G7C
 
 | | YEAST EEF1A:EEF1BA IN COMPLEX WITH GDPNP | | Descriptor: | ELONGATION FACTOR 1-ALPHA, ELONGATION FACTOR 1-BETA, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2000-11-10 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
8R61
 
 | |
6XZU
 
 | |
3EX7
 
 | |
7Q1Y
 
 | | X-ray structure of human A2ML1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin-like protein 1, ... | | Authors: | Andersen, G.R, Zarantonello, A, Enghild, J.J, Nielsen, N.S. | | Deposit date: | 2021-10-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Cryo-EM structures of human A2ML1 elucidate the protease-inhibitory mechanism of the A2M family.
Nat Commun, 13, 2022
|
|
1IJE
 
 | | Nucleotide Exchange Intermediates in the eEF1A-eEF1Ba Complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, elongation factor 1-alpha, elongation factor 1-beta | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
1IJF
 
 | | Nucleotide exchange mechanisms in the eEF1A-eEF1Ba complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, elongation factor 1-alpha, ... | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
5I8O
 
 | | HMM5 Fab in complex with disaccharide | | Descriptor: | HMM5 antibody heavy chain, HMM5 antibody light chain, alpha-L-fucopyranose-(1-3)-methyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside | | Authors: | Andersen, G.R, Tjerrild, L.M, Spillner, E. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of HMM5 Fab with a bound disaccharide
To be published
|
|
5I8K
 
 | | HHH1 Fab fragment | | Descriptor: | HHH1 Fab Heavy chain, HHH1 Fab Light chain | | Authors: | Andersen, G.R, Spillner, E. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | HHH1 Fab fragment
To be published
|
|
7ASC
 
 | | TGFBIp mutant A546T | | Descriptor: | Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Andersen, G.R, Nielsen, N.S, Gadeberg, T.A.F. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Mutation-induced dimerization of transforming growth factor-beta-induced protein may drive protein aggregation in granular corneal dystrophy.
J.Biol.Chem., 297, 2021
|
|
7AS7
 
 | |
1JKW
 
 | |
1D2E
 
 | | CRYSTAL STRUCTURE OF MITOCHONDRIAL EF-TU IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR TU (EF-TU), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Andersen, G.R, Thirup, S, Spremulli, L.L, Nyborg, J. | | Deposit date: | 1999-09-23 | | Release date: | 1999-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | High resolution crystal structure of bovine mitochondrial EF-Tu in complex with GDP.
J.Mol.Biol., 297, 2000
|
|
4F4O
 
 | | Structure of the Haptoglobin-Haemoglobin Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Andersen, C.B.F, Torvund-Jensen, M, Nielsen, M.J, Oliveira, C.L.P, Hersleth, H.P, Andersen, N.H, Pedersen, J.S, Andersen, G.R, Moestrup, S.K. | | Deposit date: | 2012-05-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the haptoglobin-haemoglobin complex.
Nature, 489, 2012
|
|
1N0V
 
 | | Crystal structure of elongation factor 2 | | Descriptor: | Elongation factor 2 | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
3KQ4
 
 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | Deposit date: | 2009-11-17 | | Release date: | 2010-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
4XW2
 
 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | Descriptor: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | Authors: | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2015-01-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
8CI4
 
 | |
