3GG2
 
 | | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate | | Descriptor: | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate
To be Published
|
|
3GDR
 
 | | Crystal structure of the D91N mutant of the orotidine 5'-monophosphate decarboxylase from Saccharomyces cerevisiae | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3GIP
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc, acetate and formate ions. | | Descriptor: | ACETIC ACID, FORMIC ACID, N-acyl-D-glutamate deacylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3G79
 
 | | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1 | | Descriptor: | NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1
To be Published
|
|
3G1F
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 5,6-dihydroorotidine 5'-monophosphate | | Descriptor: | 5,6-dihydroorotidine 5'-monophosphate, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3GDL
 
 | | Crystal structure of the orotidine 5'-monophosphate decarboxylase from Saccharomyces cerevisiae complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3GH1
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | PHOSPHATE ION, Predicted nucleotide-binding protein | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae.
To be Published
|
|
3GDO
 
 | |
3GIQ
 
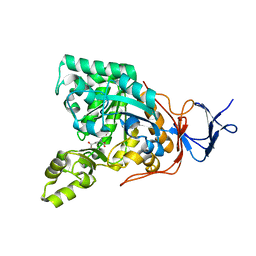 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc and phosphonate inhibitor, a mimic of the reaction tetrahedral intermediate. | | Descriptor: | N-[(R)-hydroxy(methyl)phosphoryl]-D-glutamic acid, N-acyl-D-glutamate deacylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3GFO
 
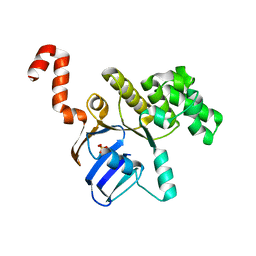 | | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ. | | Descriptor: | Cobalt import ATP-binding protein cbiO 1, SULFATE ION | | Authors: | Ramagopal, U.A, Morano, C, Toro, R, Dickey, M, Do, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ
To be published
|
|
5DM3
 
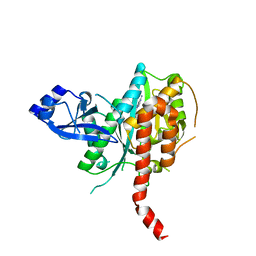 | | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-glutamine synthetase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP
To be published
|
|
5E5M
 
 | | Crystal structure of mouse CTLA-4 in complex with nanobody | | Descriptor: | CTLA-4 nanobody, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-12 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Crystal structure of mouse CTLA-4 in complex with nanobody
To Be Published
|
|
5DXW
 
 | | Crystal structure of mouse PD-L1 nanobody | | Descriptor: | PD-L1 nanobody, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Almo, S.C, Ploegh, H, Ingram, J, Dougan, M. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Crystal structure of mouse PD-L1 nanobody
To Be Published
|
|
5E03
 
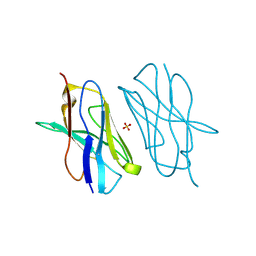 | | Crystal structure of mouse CTLA-4 nanobody | | Descriptor: | CTLA-4 nanobody, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Crystal structure of mouse CTLA-4 nanobody
To Be Published
|
|
5ESR
 
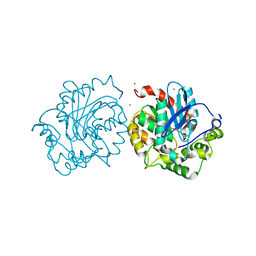 | | Crystal structure of haloalkane dehalogenase (DccA) from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Haloalkane dehalogenase, ... | | Authors: | Malashkevich, V.N, Toro, R, Mundorff, E.C, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Biochemical characterization of two haloalkane dehalogenases: DccA from Caulobacter crescentus and DsaA from Saccharomonospora azurea.
Protein Sci., 25, 2016
|
|
5DMH
 
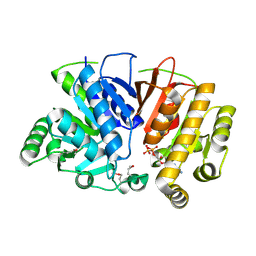 | | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein conserved in bacteria | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
To be published
|
|
5E83
 
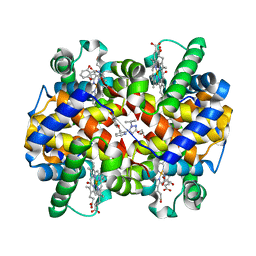 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT440, CO-CRYSTALLIZATION EXPERIMENT | | Descriptor: | 2-methyl-3-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)phenol, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Patskovska, L, Patskovsky, Y, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GBT440 increases haemoglobin oxygen affinity, reduces sickling and prolongs RBC half-life in a murine model of sickle cell disease.
Br.J.Haematol., 175, 2016
|
|
5DTE
 
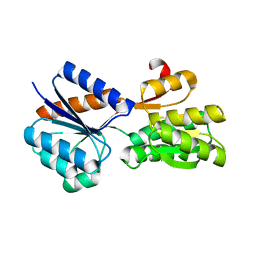 | | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose | | Descriptor: | Monosaccharide-transporting ATPase, beta-D-allopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose
To be published
|
|
5E56
 
 | | Crystal structure of mouse CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, SAMANTA, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Crystal structure of mouse CTLA-4
To Be Published
|
|
5DKV
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose | | Descriptor: | ABC transporter substrate binding protein (Ribose), alpha-D-tagatopyranose | | Authors: | Yadava, U, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose
To be published
|
|
4WCZ
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
4WK5
 
 | | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458 | | Descriptor: | Geranyltranstransferase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458
To be published
|
|
4WGH
 
 | | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution | | Descriptor: | ACETATE ION, Aldehyde reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Hillerich, B.S, Zimmerman, M.D, Chowdhury, S, Hammonds, J, Al Obaidi, N, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution
to be published
|
|
4WKC
 
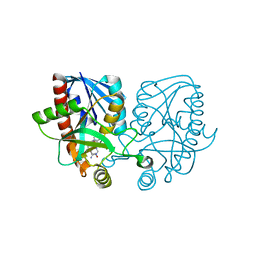 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TETRAETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
4WUT
 
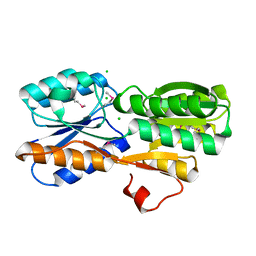 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
