5CRM
 
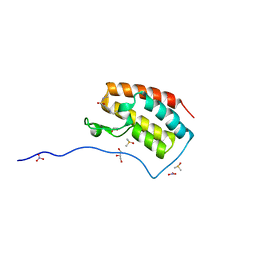 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CQT
 
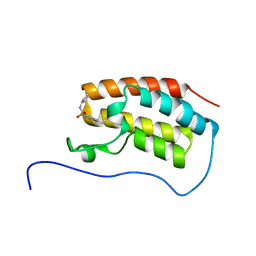 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-cyclohexyl-1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CY9
 
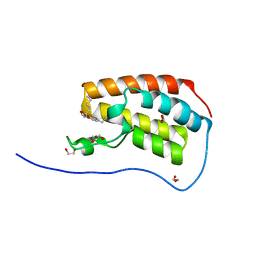 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-30 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CRZ
 
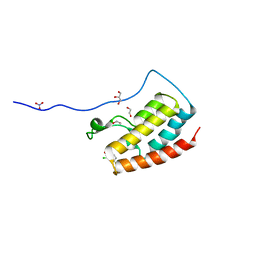 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-4-fluorobenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
6IUP
 
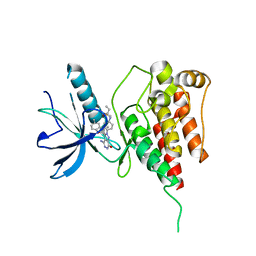 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
5DX4
 
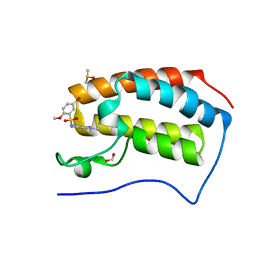 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-2-methoxybenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5FDP
 
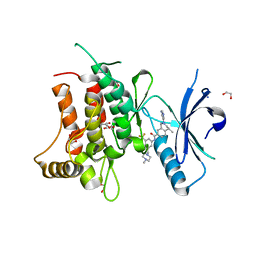 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
6BVY
 
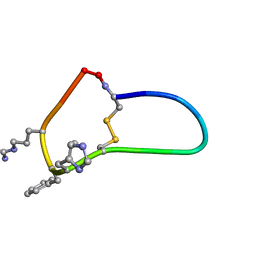 | | SFTI-HFRW-4 | | Descriptor: | Trypsin inhibitor 1 HFRW-4 | | Authors: | Schroeder, C.I, White, A. | | Deposit date: | 2017-12-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J. Med. Chem., 61, 2018
|
|
3PPV
 
 | | Crystal structure of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | CALCIUM ION, SULFATE ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPW
 
 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPY
 
 | | Crystal structure of the D1596A/N1602A double mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
5DG1
 
 | | Sugar binding protein - human galectin-2 | | Descriptor: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim.Biophys.Sin., 2016
|
|
5IMX
 
 | |
5DG2
 
 | | Sugar binding protein - human galectin-2 (dimer) | | Descriptor: | Galectin-2, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim. Biophys. Sin. (Shanghai), 48, 2016
|
|
5VRN
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-4-[[5-[4-cyano-2-[(~{E})-hydroxyiminomethyl]phenoxy]-1-oxidanyl-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5VRM
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3-oxidanyl-4-[[1-oxidanyl-6-[4-(trifluoromethyl)phenoxy]-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5VRL
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | (~{N}~{E})-~{N}-[[2-[[2-ethylsulfonyl-1,1-bis(oxidanyl)-3,4-dihydro-2,3,1$l^{4}-benzodiazaborinin-7-yl]oxy]-5-(trifluoromethyl)phenyl]methylidene]hydroxylamine, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5EWS
 
 | | Sugar binding protein - human galectin-2 | | Descriptor: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2015-11-21 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim. Biophys. Sin. (Shanghai), 48, 2016
|
|
3CVR
 
 | | Crystal structure of the full length IpaH3 | | Descriptor: | Invasion plasmid antigen | | Authors: | Zhu, Y, Shao, F. | | Deposit date: | 2008-04-19 | | Release date: | 2008-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Shigella effector reveals a new class of ubiquitin ligases
Nat.Struct.Mol.Biol., 15, 2008
|
|
6S4Q
 
 | | scdSav(SASK) - Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes | | Descriptor: | GLYCEROL, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-06-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Breaking Symmetry: Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes.
J.Am.Chem.Soc., 141, 2019
|
|
8J40
 
 | | Crystal Structure of CATB8 in complex with chloramphenicol | | Descriptor: | CHLORAMPHENICOL, CatB8, GLYCEROL, ... | | Authors: | Liao, J, Kuang, L, Jiang, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Chloramphenicol Binding Sites of Acinetobacter baumannii Chloramphenicol Acetyltransferase CatB8.
Acs Infect Dis., 10, 2024
|
|
6S50
 
 | |
6T38
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6T37
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
