6WJY
 
 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4-A | | Descriptor: | 3-chloro-N-(3-{(2S)-1-[(4-fluorophenyl)amino]-1-oxopropan-2-yl}bicyclo[1.1.1]pentan-1-yl)benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A, Neumann, L. | | Deposit date: | 2020-04-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of Potent and Orally Available Bicyclo[1.1.1]pentane-Derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
3KMC
 
 | | Crystal structure of catalytic domain of TACE with tartrate-based inhibitor | | Descriptor: | (2R,3R)-4-[4-(2-chlorophenyl)piperazin-1-yl]-2,3-dihydroxy-4-oxo-N-(2-thiophen-2-ylethyl)butanamide, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, TNF-alpha-converting enzyme, ... | | Authors: | Orth, P. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The discovery of novel tartrate-based TNF-alpha converting enzyme (TACE) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2009
|
|
6PZE
 
 | |
6PZF
 
 | |
6PZG
 
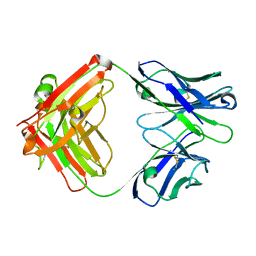 | | Crystal structure of human NA-80 Fab | | Descriptor: | NA-80 Fab heavy chain, NA-80 Fab light chain | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6Q23
 
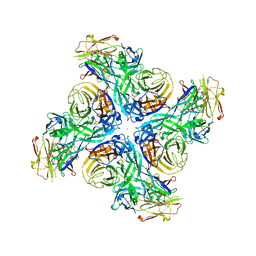 | |
6PZZ
 
 | | CryoEM derived model of NA-80 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-80 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6PZH
 
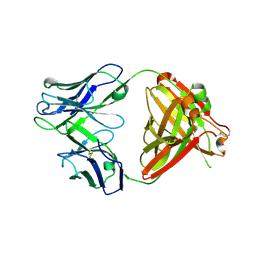 | | Crystal structure of human NA-22 Fab | | Descriptor: | NA-22 Fab heavy chain, NA-22 Fab light chain | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6Q20
 
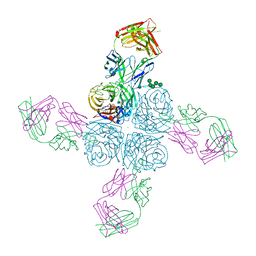 | |
6PU7
 
 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
6PZY
 
 | | CryoEM derived model of NA-73 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-73 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
4F3Z
 
 | | Crystal structure of a swine H1N2 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Functional Balance of the Hemagglutinin and Neuraminidase Activities Accompanies the Emergence of the 2009 H1N1 Influenza Pandemic.
J.Virol., 86, 2012
|
|
4O58
 
 | |
4O5N
 
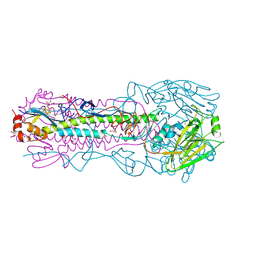 | | Crystal structure of A/Victoria/361/2011 (H3N2) influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.
Nat Commun, 5, 2014
|
|
4O5L
 
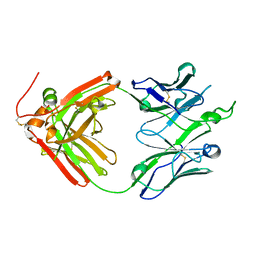 | | Crystal structure of broadly neutralizing antibody F045-092 | | Descriptor: | Fab F045-092 heavy chain, Fab F045-092 light chain, PHOSPHATE ION | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5045 Å) | | Cite: | Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.
Nat Commun, 5, 2014
|
|
4O5I
 
 | |
6X3N
 
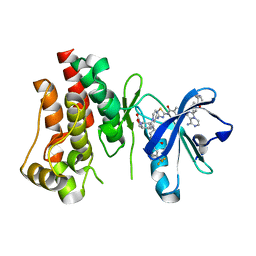 | | Co-structure of BTK kinase domain with L-005085737 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxohexahydro-3H-[1,3]oxazolo[3,4-a]pyridin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X3O
 
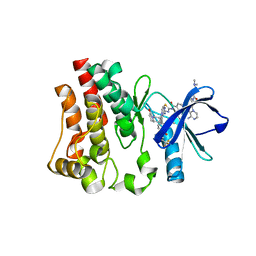 | | Co-structure of BTK kinase domain with L-005191930 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxo-3,5,6,7,8,8a-hexahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-methoxy-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
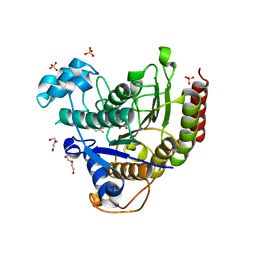
6XDM
 
 | |
7U2E
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADI-55688 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-55688 heavy chain, ADI-55688 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U2D
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADG20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADG20 heavy chain, ADG20 light chain, ... | | Authors: | Zhu, X, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC2
 
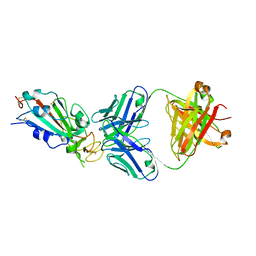 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC3
 
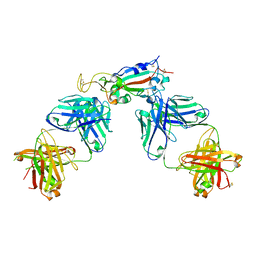 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.1 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
