5JQG
 
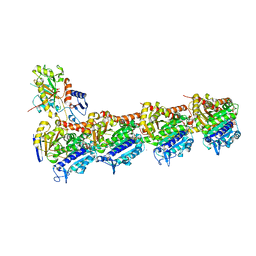 | | An apo tubulin-RB-TTL complex structure used for side-by-side comparison | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Naismith, J.H, Zhu, X. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule
Nat Commun, 7, 2016
|
|
8KFA
 
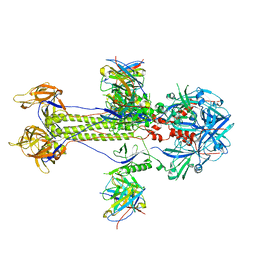 | | Cryo-EM structure of HSV-1 gB with D48 Fab complex | | Descriptor: | D48 heavy chain, D48 light chain, Envelope glycoprotein B | | Authors: | Yang, J, Sun, C, Fang, X, Zeng, M, Liu, Z. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | The structure of HSV-1 gB bound to a potent neutralizing antibody reveals a conservative antigenic domain across herpesviruses
hlife, 2023
|
|
5IFG
 
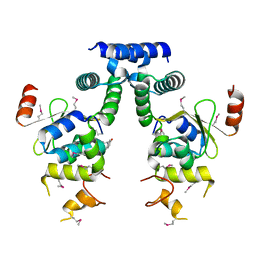 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
7TGL
 
 | |
7TGK
 
 | | Crystal structure of ATP bound DesD, the desferrioxamine synthetase from the Streptomyces griseoflavus ferrimycin biosynthetic pathway | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
7TGN
 
 | | Crystal structure of DesD, the desferrioxamine synthetase from the Streptomyces violaceus salmycin biosynthetic pathway | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Desferrioxamine synthetase DesD, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
7TGM
 
 | |
7TGJ
 
 | |
7CE8
 
 | | Crystal structure of T2R-TTL-Compound11 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
8TGP
 
 | |
7CDA
 
 | | Crystal structure of T2R-TTL-PAC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CEK
 
 | | Crystal structure of T2R-TTL-BML-284 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CE6
 
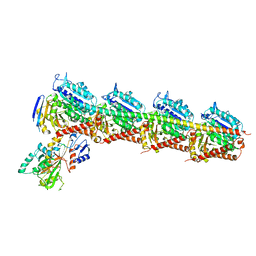 | | Crystal structure of T2R-TTL-Compound9 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
1WAO
 
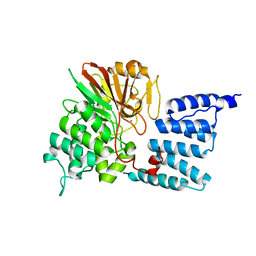 | | PP5 structure | | Descriptor: | MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Barford, D. | | Deposit date: | 2004-10-27 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for Tpr Domain-Mediated Regulation of Protein Phosphatase 5
Embo J., 24, 2005
|
|
3G6L
 
 | |
6JRX
 
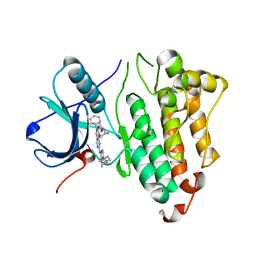 | | EGFR T790M/C797S in complex with compound 6i | | Descriptor: | Epidermal growth factor receptor, N-{trans-4-[3-(2-chlorophenyl)-7-{[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]cyclohexyl}propanamide | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
5YDG
 
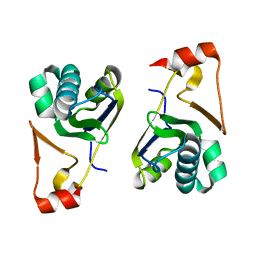 | | Crystal structure of the Arabidopsis thaliana chloroplast RNA editing factors 2(MORF2) | | Descriptor: | Multiple organellar RNA editing factor 2, chloroplastic | | Authors: | Wang, X, Yang, J.Y, Wang, Y.L, Gao, Y.S. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of the chloroplast RNA editing factor MORF2
Biochem. Biophys. Res. Commun., 495, 2018
|
|
6JUU
 
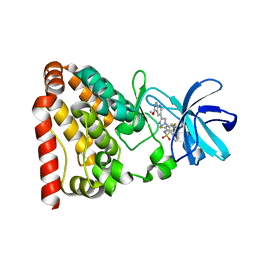 | | Crystal structure of ZAK in complex with compound 6r | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]naphthalene-1-sulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JUT
 
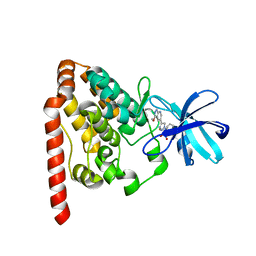 | | Crystal structure of ZAK in complex with compound 6k | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
2JM1
 
 | |
6AH7
 
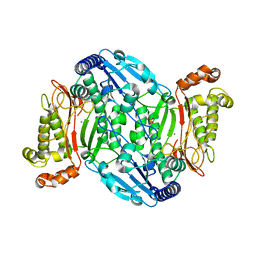 | | D45W/H226G mutant of marine bacterial prolidase | | Descriptor: | MANGANESE (II) ION, SODIUM ION, SULFATE ION, ... | | Authors: | Jian, Y, Yunzhu, X, Lijuan, L. | | Deposit date: | 2018-08-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Repurposing a bacterial prolidase for organophosphorus hydrolysis: Reshaped catalytic cavity switches substrate selectivity.
Biotechnol.Bioeng., 117, 2020
|
|
6AH8
 
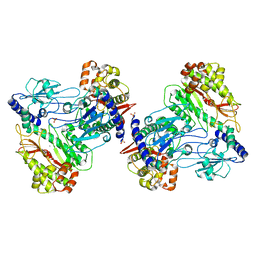 | |
7WRX
 
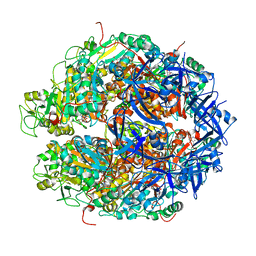 | | Structure of Deinococcus radiodurans HerA-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HerA, MAGNESIUM ION | | Authors: | Cheng, K. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.40003562 Å) | | Cite: | Structural and DNA end resection study of the bacterial NurA-HerA complex.
Bmc Biol., 21, 2023
|
|
7WRW
 
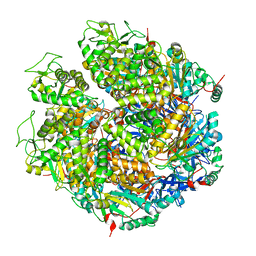 | | Structure of Deinococcus radiodurans HerA | | Descriptor: | HerA | | Authors: | Cheng, K. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.00008273 Å) | | Cite: | Structural and DNA end resection study of the bacterial NurA-HerA complex.
Bmc Biol., 21, 2023
|
|
6YYE
 
 | | TREM2 extracellular domain (19-131) in complex with single-chain variable fragment (scFv-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TREM2 Single chain variable 2, Triggering receptor expressed on myeloid cells 2 | | Authors: | Szykowska, A, Preger, C, Krojer, T, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-05-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
