4WD1
 
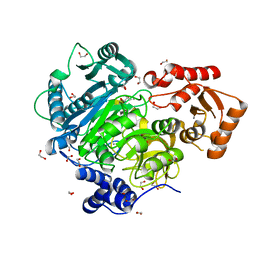 | | Acetoacetyl-CoA Synthetase from Streptomyces lividans | | Descriptor: | 1,2-ETHANEDIOL, Acetoacetate-CoA ligase, CALCIUM ION | | Authors: | Gulick, A.M, Mitchell, C.A. | | Deposit date: | 2014-09-05 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The structure of S. lividans acetoacetyl-CoA synthetase shows a novel interaction between the C-terminal extension and the N-terminal domain.
Proteins, 83, 2015
|
|
4RCV
 
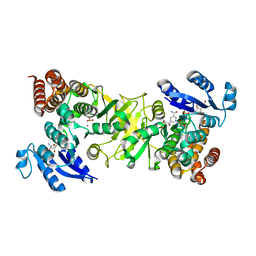 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase bound to 1-deoxy-L-erythrulose | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 1-deoxy-L-erythrulose, MANGANESE (II) ION, ... | | Authors: | Gulick, A.M, Allen, C.L, Kholodar, S.A, Murkin, A.S. | | Deposit date: | 2014-09-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | The role of phosphate in a multistep enzymatic reaction: reactions of the substrate and intermediate in pieces.
J.Am.Chem.Soc., 137, 2015
|
|
6MFL
 
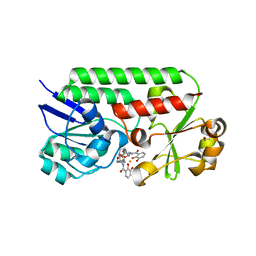 | | Structure of siderophore binding protein BauB bound to a complex between two molecules of acinetobactin and ferric iron. | | Descriptor: | 1,2-ETHANEDIOL, BauB, FE (III) ION, ... | | Authors: | Gulick, A.M, Bailey, D.C, Wencewicz, T.A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Siderophore Binding Protein BauB Bound to an Unusual 2:1 Complex Between Acinetobactin and Ferric Iron.
Biochemistry, 57, 2018
|
|
1T5D
 
 | |
1T5H
 
 | |
6PYP
 
 | | Binary Complex of Human Glycerol 3-Phosphate Dehydrogenase, R269A mutant | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic, ... | | Authors: | Gulick, A.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modeling the Role of a Flexible Loop and Active Site Side Chains in Hydride Transfer Catalyzed by Glycerol-3-phosphate Dehydrogenase.
Acs Catalysis, 10, 2020
|
|
1PG3
 
 | | Acetyl CoA Synthetase, Acetylated on Lys609 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, COENZYME A, ... | | Authors: | Gulick, A.M, Starai, V.J, Horswill, A.R, Homick, K.M, Escalante-Semerena, J.C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.75 A Crystal Structure of Acetyl-CoA Synthetase Bound
to Adenosine-5'-propylphosphate and Coenzyme A
Biochemistry, 42, 2003
|
|
1PG4
 
 | | Acetyl CoA Synthetase, Salmonella enterica | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, CHLORIDE ION, ... | | Authors: | Gulick, A.M, Starai, V.J, Horswill, A.R, Homick, K.M, Escalante-Semerena, J.C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.75 A Crystal Structure of Acetyl-CoA Synthetase Bound
to Adenosine-5'-propylphosphate and Coenzyme A
Biochemistry, 42, 2003
|
|
2FWM
 
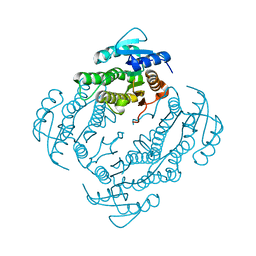 | | Crystal Structure of E. coli EntA, a 2,3-dihydrodihydroxy benzoate dehydrogenase | | Descriptor: | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | | Authors: | Gulick, A.M, Duax, W.L. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of the crystal structure of EntA, a 2,3-dihydro-2,3-dihydroxybenzoic acid dehydrogenase from Escherichia coli.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5KYV
 
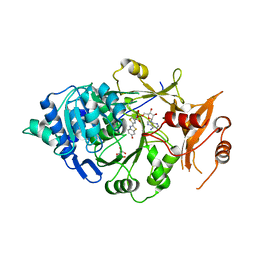 | | Structure of Photinus pyralis Luciferase green shifted light emitting variant | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, L(+)-TARTARIC ACID, Luciferin 4-monooxygenase | | Authors: | Gulick, A.M. | | Deposit date: | 2016-07-22 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cloning of the Orange Light-Producing Luciferase from Photinus scintillans Provides Insight into Bioluminescence Color Determination
To Be Published
|
|
5KYT
 
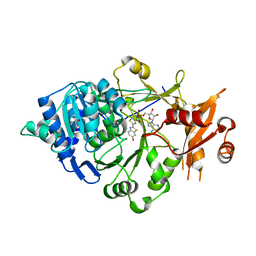 | | Structure of Photinus pyralis Luciferase red light emitting variant | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, Luciferin 4-monooxygenase | | Authors: | Gulick, A.M. | | Deposit date: | 2016-07-22 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Cloning of the Orange Light-Producing Luciferase from Photinus scintillans-A New Proposal on how Bioluminescence Color is Determined.
Photochem. Photobiol., 93, 2017
|
|
1JCT
 
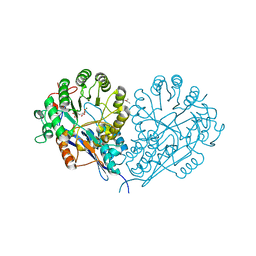 | | Glucarate Dehydratase, N341L mutant Orthorhombic Form | | Descriptor: | D-GLUCARATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-11 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1JDF
 
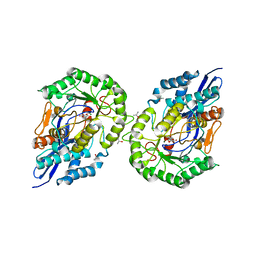 | | Glucarate Dehydratase from E.coli N341D mutant | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-13 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
3GNM
 
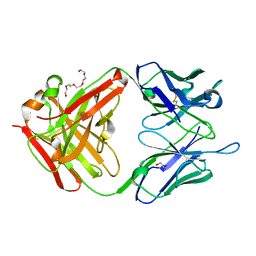 | | The crystal structure of the JAA-F11 monoclonal antibody Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, JAA-F11 Fab Antibody Fragment, Heavy Chain, ... | | Authors: | Gulick, A.M, Rittenhouse-Olson, K, Jadey, S. | | Deposit date: | 2009-03-17 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational Screening of the Human TF-Glycome Provides a Structural Definition for the Specificity of Anti-Tumor Antibody JAA-F11.
Plos One, 8, 2013
|
|
3KAR
 
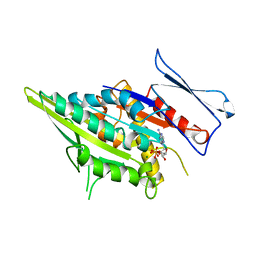 | | THE MOTOR DOMAIN OF KINESIN-LIKE PROTEIN KAR3, A SACCHAROMYCES CEREVISIAE KINESIN-RELATED PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Gulick, A.M, Song, H, Endow, S, Rayment, I. | | Deposit date: | 1997-11-26 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the yeast Kar3 motor domain complexed with Mg.ADP to 2.3 A resolution.
Biochemistry, 37, 1998
|
|
1EC7
 
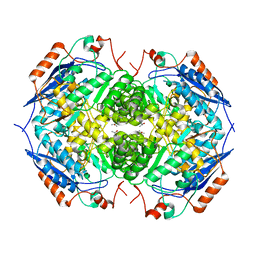 | | E. COLI GLUCARATE DEHYDRATASE NATIVE ENZYME | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1ECQ
 
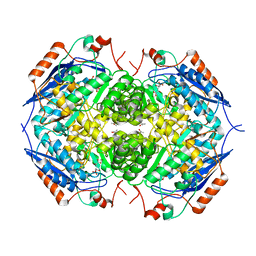 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO 4-DEOXYGLUCARATE | | Descriptor: | 4-DEOXYGLUCARATE, GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EC8
 
 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO PRODUCT 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EC9
 
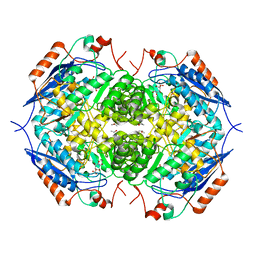 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO XYLAROHYDROXAMATE | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
3SRC
 
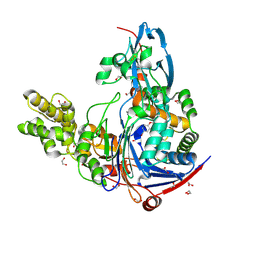 | | Structure of Pseudomonas aeruginosa PvdQ bound to NS2028 | | Descriptor: | 1,2-ETHANEDIOL, 8-bromo-4H-[1,2,4]oxadiazolo[3,4-c][1,4]benzoxazin-1-one, Acyl-homoserine lactone acylase pvdQ | | Authors: | Gulick, A.M, Drake, E.J. | | Deposit date: | 2011-07-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis.
Acs Chem.Biol., 6, 2011
|
|
3SRA
 
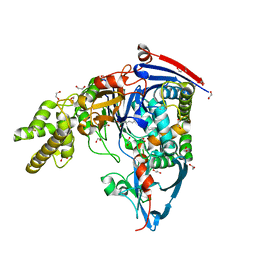 | | Structure of Pseudomonas aerugionsa PvdQ covalently acylated with myristic acid from PVDIq | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, ... | | Authors: | Gulick, A.M, Drake, E.J. | | Deposit date: | 2011-07-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis.
Acs Chem.Biol., 6, 2011
|
|
3SRB
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to SMER28 | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-N-(prop-2-en-1-yl)quinazolin-4-amine, Acyl-homoserine lactone acylase pvdQ, ... | | Authors: | Gulick, A.M, Drake, E.J. | | Deposit date: | 2011-07-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis.
Acs Chem.Biol., 6, 2011
|
|
3U17
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-(p-benzoyl)phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(4-benzoylphenyl)-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Gulick, A.M, Drake, E.J, Aldrich, C.C, Neres, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii.
J.Med.Chem., 56, 2013
|
|
3U16
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-(p-benzyloxy)phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-(benzyloxy)phenyl]-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Gulick, A.M, Drake, E.J, Aldrich, C.C, Neres, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii.
J.Med.Chem., 56, 2013
|
|
4IZ6
 
 | | Structure of EntE and EntB, an NRPS adenylation-PCP fusion protein with pseudo translational symmetry | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-deoxy-5'-({[2-(2,3-dihydroxyphenyl)ethyl]sulfonyl}amino)adenosine, Enterobactin synthase component E, ... | | Authors: | Gulick, A.M, Sundlov, J.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure determination of the functional domain interaction of a chimeric nonribosomal peptide synthetase from a challenging crystal with noncrystallographic translational symmetry.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
