5WS6
 
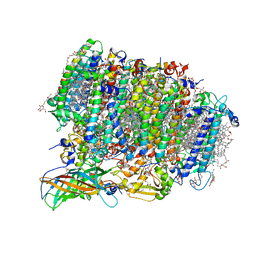 | | Native XFEL structure of Photosystem II (preflash two-flash dataset | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
3G5S
 
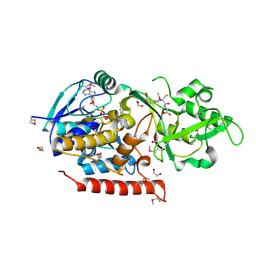 | | Crystal structure of Thermus thermophilus TrmFO in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5Q
 
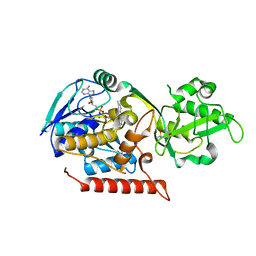 | | Crystal structure of Thermus thermophilus TrmFO | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5R
 
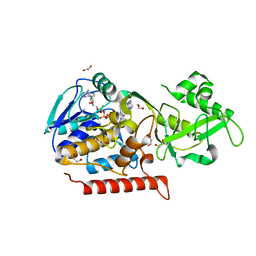 | | Crystal structure of Thermus thermophilus TrmFO in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6GTL
 
 | | Achromobacter cycloclastes copper nitrite reductase at pH 6.0 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GTJ
 
 | | Neutron crystal structure for copper nitrite reductase from Achromobacter Cycloclastes at 1.8 A resolution | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Antonyuk, S.V, Blakeley, M.P, Halsted, T.P, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | NEUTRON DIFFRACTION (1.801 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GTI
 
 | | Achromobacter cycloclastes copper nitrite reductase at pH 5.0 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GTK
 
 | | Achromobacter cycloclastes copper nitrite reductase at pH 5.5 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GTN
 
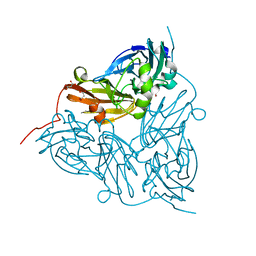 | | Achromobacter cycloclastes copper nitrite reductase at pH 6.5 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
2D7T
 
 | |
2DUO
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Vesicular integral-membrane protein VIP36 | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUR
 
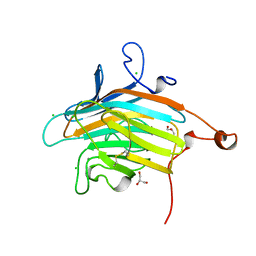 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man2-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUQ
 
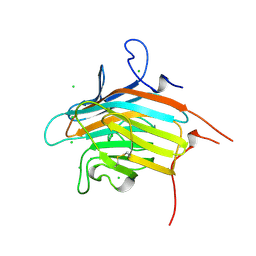 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Vesicular integral-membrane protein VIP36, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUP
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, metal-free form | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2E6V
 
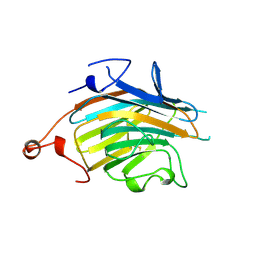 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man3GlcNAc-bound form | | Descriptor: | CALCIUM ION, Vesicular integral-membrane protein VIP36, alpha-D-mannopyranose, ... | | Authors: | Satoh, T, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
8P3U
 
 | |
5GKR
 
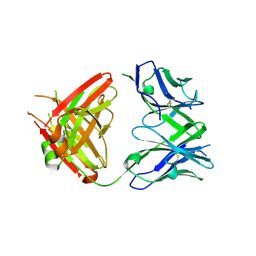 | | Crystal structure of SLE patient-derived anti-DNA antibody in complex with oligonucleotide | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), IgG2, Fab (heavy chain), ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
5GKS
 
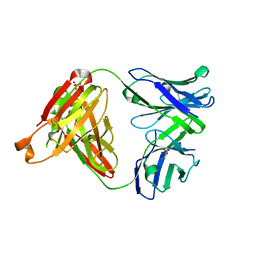 | | Crystal structure of SLE patient-derived anti-DNA antibody | | Descriptor: | IgG2, Fab (heavy chain), PHOSPHATE ION, ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
5B4Y
 
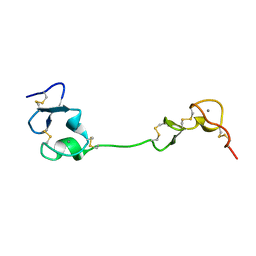 | | Crystal structure of the LA12 fragment of ApoER2 | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 8 | | Authors: | Nogi, T, Tabata, S, Hirai, H, Yasui, N, Takagi, J. | | Deposit date: | 2016-04-20 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
To Be Published
|
|
7EA6
 
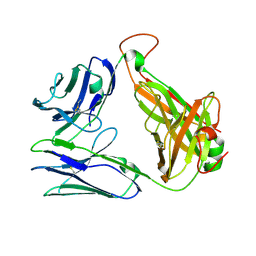 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
7VI4
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, A381T mutant | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.95 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VI5
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, wild type sequence | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.761 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5YFI
 
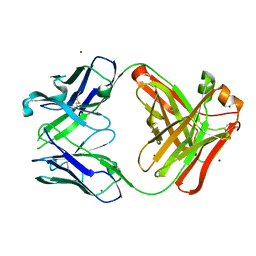 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-21 | | Release date: | 2018-12-05 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YHL
 
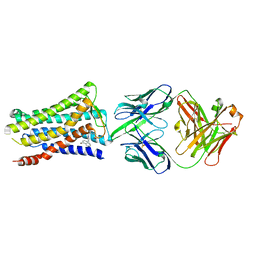 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | Descriptor: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-28 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YWY
 
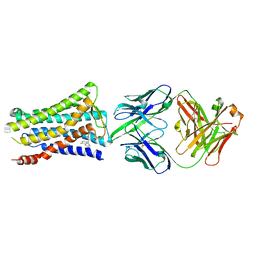 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | Descriptor: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
