6G9B
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with imipramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, ... | | Authors: | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
6G95
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
6G9I
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with clomipramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(3-CHLORO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, Envelope glycoprotein,Envelope glycoprotein, ... | | Authors: | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
7EYA
 
 | |
7EY0
 
 | |
7E7Y
 
 | |
7E88
 
 | |
7E7X
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
7Y0C
 
 | | Crystal structure of BD55-1403 and SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-1403 Fab heavy chain, BD55-1403 Fab light chain, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7Y0W
 
 | | Local structure of BD55-5514 and BD55-5840 Fab and Omicron BA.1 RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-5514H, BD55-5514L, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
4GY5
 
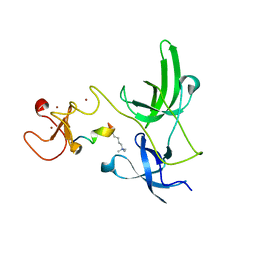 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
7V20
 
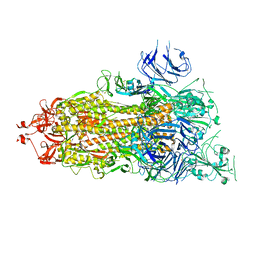 | | CryoEM structure of del68-76/del679-688 prefusion-stabilized spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Du, S, Xiao, J. | | Deposit date: | 2021-08-07 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | CryoEM structure of del68-76/del679-688 prefusion-stabilized spike
To Be Published
|
|
7V24
 
 | |
7V22
 
 | |
7V23
 
 | |
1PGY
 
 | | Solution structure of the UBA domain in Saccharomyces cerevisiae protein, Swa2p | | Descriptor: | Swa2p | | Authors: | Chim, N, Gall, W.E, Xiao, J, Harris, M.P, Graham, T.R, Krezel, A.M. | | Deposit date: | 2003-05-28 | | Release date: | 2004-03-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-binding domain in Swa2p from Saccharomyces cerevisiae.
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
5X83
 
 | | Structure of DCC FN456 domains | | Descriptor: | Netrin receptor DCC | | Authors: | Finci, F.I, Xiao, J, Wang, J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structure of unliganded membrane-proximal domains FN4-FN5-FN6 of DCC
Protein Cell, 8, 2017
|
|
5ZQZ
 
 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZRV
 
 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XOO
 
 | | The structure of hydra Fam20 with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE, Glycosaminoglycan xylosylkinase, ... | | Authors: | Zhang, H, Xiao, J. | | Deposit date: | 2017-05-29 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
5YH3
 
 | | The structure of hFam20C and hFam20A complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Extracellular serine/threonine protein kinase FAM20C, Pseudokinase FAM20A | | Authors: | Zhu, Q, Xiao, J. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
5YH0
 
 | | The structure of DrFam20C1 | | Descriptor: | DrFam20C1 | | Authors: | Zhang, H, Xiao, J. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
