3RHX
 
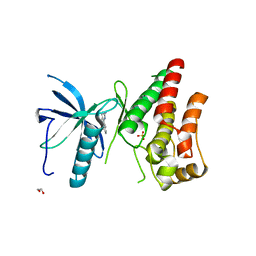 | | Crystal structure of the catalytic domain of FGFR1 kinase in complex with ARQ 069 | | Descriptor: | (6S)-6-phenyl-5,6-dihydrobenzo[h]quinazolin-2-amine, 1,2-ETHANEDIOL, Basic fibroblast growth factor receptor 1, ... | | Authors: | Eathiraj, S, Palma, R, Hirschi, M, Volckova, E, Nakuci, E, Castro, J, Chen, C.R, Chan, T.C, France, D.S, Ashwell, M.A. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.
J.Biol.Chem., 286, 2011
|
|
3RI1
 
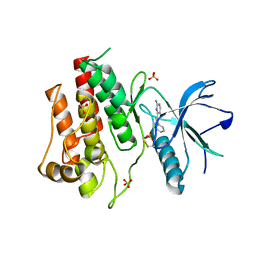 | | Crystal structure of the catalytic domain of FGFR2 kinase in complex with ARQ 069 | | Descriptor: | (6S)-6-phenyl-5,6-dihydrobenzo[h]quinazolin-2-amine, Fibroblast growth factor receptor 2, SULFATE ION | | Authors: | Eathiraj, S, Palma, R, Hirschi, M, Volckova, E, Nakuci, E, Castro, J, Chen, C.R, Chan, T.C, France, D.S, Ashwell, M.A. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-04 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.
J.Biol.Chem., 286, 2011
|
|
3RHK
 
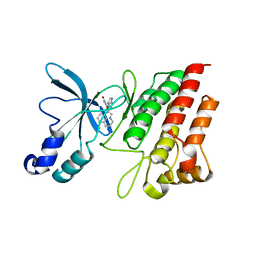 | | Crystal structure of the catalytic domain of c-Met kinase in complex with ARQ 197 | | Descriptor: | 1-[(3R,4R)-4-(1H-indol-3-yl)-2,5-dioxopyrrolidin-3-yl]pyrrolo[3,2,1-ij]quinolinium, Hepatocyte growth factor receptor | | Authors: | Eathiraj, S, Palma, R, Volckova, E, Hirschi, M, France, D.S, Ashwell, M.A, Chan, T.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197.
J.Biol.Chem., 286, 2011
|
|
4EJN
 
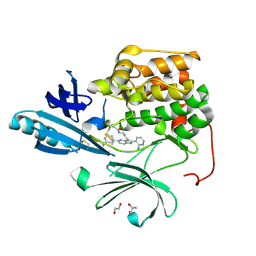 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
5KCV
 
 | | Crystal structure of allosteric inhibitor, ARQ 092, in complex with autoinhibited form of AKT1 | | Descriptor: | 3-[3-[4-(1-azanylcyclobutyl)phenyl]-5-phenyl-imidazo[4,5-b]pyridin-2-yl]pyridin-2-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Eathiraj, S. | | Deposit date: | 2016-06-07 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
J.Med.Chem., 59, 2016
|
|
1K0P
 
 | |
1K18
 
 | |
