4K1G
 
 | |
8CKO
 
 | | PBP AccA from A.tumefaciens C58 in complex with agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate-binding protein, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CKE
 
 | |
6EPY
 
 | |
6QAK
 
 | |
6QAP
 
 | |
6QAO
 
 | |
3RX5
 
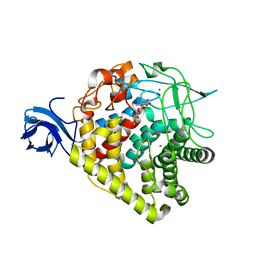 | | structure of AaCel9A in complex with cellotriose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl 4-O-beta-D-glucopyranosyl-beta-D-glucopyranoside, CALCIUM ION, Cellulase, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A fortuitous binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidase
Org.Biomol.Chem., 9, 2011
|
|
4I8P
 
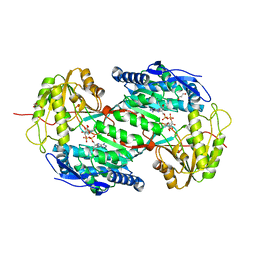 | | Crystal structure of aminoaldehyde dehydrogenase 1a from Zea mays (ZmAMADH1a) | | Descriptor: | 1,2-ETHANEDIOL, Aminoaldehyde dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4I8Q
 
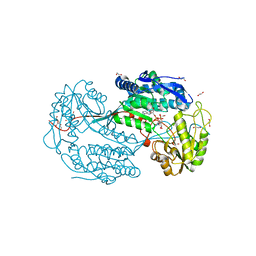 | | Structure of the aminoaldehyde dehydrogenase 1 E260A mutant from Solanum lycopersicum (SlAMADH1-E260A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4I9B
 
 | | Structure of aminoaldehyde dehydrogenase 1 from Solanum lycopersium (SlAMADH1) with a thiohemiacetal intermediate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
5LOM
 
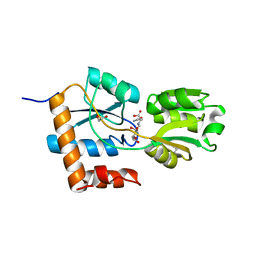 | | Crystal structure of the PBP SocA from Agrobacterium tumefaciens C58 in complex with DFG at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Deoxyfructosyl-amino Acid Transporter Periplasmic Binding Protein, Deoxyfructosylglutamine | | Authors: | Marty, L, Vigouroux, A, Morera, S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
4KPN
 
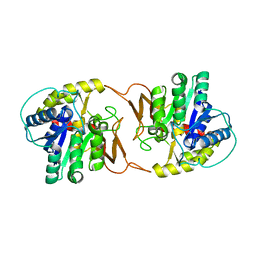 | | Plant nucleoside hydrolase - PpNRh1 enzyme | | Descriptor: | CALCIUM ION, Nucleoside N-ribohydrolase 1 | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure and Function of Nucleoside Hydrolases from Physcomitrella patens and Maize Catalyzing the Hydrolysis of Purine, Pyrimidine, and Cytokinin Ribosides.
Plant Physiol., 163, 2013
|
|
4KPO
 
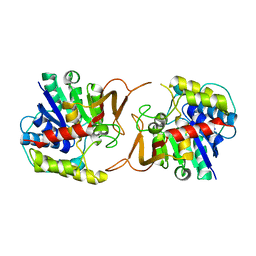 | | Plant nucleoside hydrolase - ZmNRh3 enzyme | | Descriptor: | CALCIUM ION, Nucleoside N-ribohydrolase 3 | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and Function of Nucleoside Hydrolases from Physcomitrella patens and Maize Catalyzing the Hydrolysis of Purine, Pyrimidine, and Cytokinin Ribosides.
Plant Physiol., 163, 2013
|
|
5MZ8
 
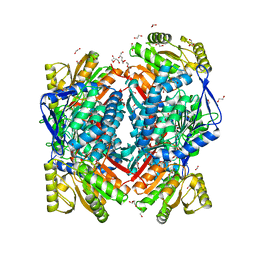 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
3IP5
 
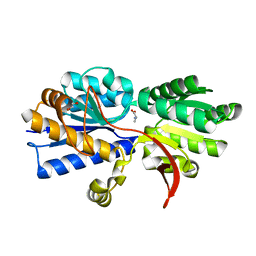 | | Structure of Atu2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP7
 
 | | Structure of Atu2422-GABA receptor in complex with valine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), CALCIUM ION, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP6
 
 | | Structure of Atu2422-GABA receptor in complex with proline | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), PROLINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IPC
 
 | | Structure of ATU2422-GABA F77A mutant receptor in complex with leucine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), LEUCINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IPA
 
 | | Structure of ATU2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP9
 
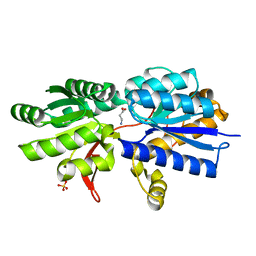 | | Structure of Atu2422-GABA receptor in complex with GABA | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
4E9E
 
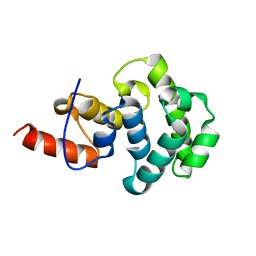 | | Structure of the glycosylase domain of MBD4 | | Descriptor: | Methyl-CpG-binding domain protein 4 | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2012-03-21 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
4E9H
 
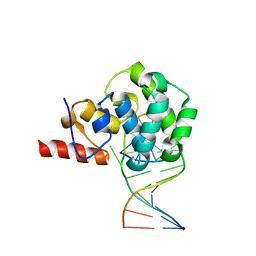 | | structure of glycosylase domain of MBD4 bound to 5hmU containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(5HU)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2012-03-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
4EA5
 
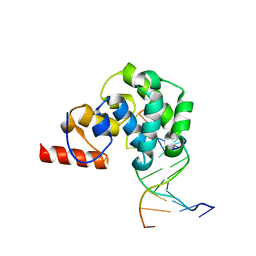 | | Structure of the glycoslyase domain of MBD4 bound to a 5hmU containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(5HU)*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2012-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
4E9G
 
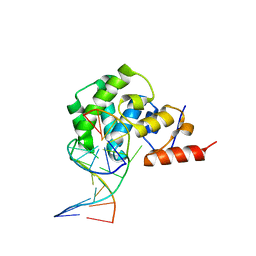 | | structure of the glycosylase domain of MBD4 bound to thymine containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2012-03-21 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
