3FX8
 
 | | Distinct recognition of three-way DNA junctions by a thioester variant of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | (5'-D(*CP*GP*TP*AP*CP*G)-3', 4,4'-sulfanediylbis{N-[(1E)-pyridin-2-ylmethylidene]aniline}, FE (II) ION | | Authors: | Boer, D.R, Uson, I, Coll, M. | | Deposit date: | 2009-01-20 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3GNG
 
 | | P21B crystal structure of R1-R7 of Murine MVP | | Descriptor: | Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Uson, I, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
3GNF
 
 | | P1 Crystal structure of the N-terminal R1-R7 of murine MVP | | Descriptor: | Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Uson, I, Luque, D, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
3HCL
 
 | | Helical superstructures in a DNA oligonucleotide crystal | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*AP*T)-3') | | Authors: | De Luchi, D, Martinez de Ilarduya, I, Subirana, J.A, Uson, I, Campos, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A geometric approach to the crystallographic solution of nonconventional DNA structures: helical superstructures of d(CGATAT)
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7QFI
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain I (aa 31-182) | | Descriptor: | CALCIUM ION, SlpX | | Authors: | Sagmeister, T, Damisch, E, Millan, C, Uson, I, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The self-assembly of the S-layer protein from Lactobacilli acidophilus
To be published
|
|
7QEC
 
 | | Crystal structure of SlpA - domain II, domain that is involved in the self-assembly of the S-layer from Lactobacillus amylovorus | | Descriptor: | S-layer | | Authors: | Eder, M, Dordic, A, Millan, C, Sagmeister, T, Uson, I, Pavkov-Keller, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The self-assembly of the S-layer protein from lactobacilli
to be published
|
|
7TA2
 
 | | Crystal structure of the human sperm-expressed surface protein SPACA6 | | Descriptor: | BROMIDE ION, Sperm acrosome membrane-associated protein 6 | | Authors: | Vance, T.D.R, Yip, P, Jimenez, E, Uson, I, Lee, J.E. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins.
Commun Biol, 5, 2022
|
|
6X8O
 
 | | BimBH3 peptide tetramer | | Descriptor: | Bcl-2-like protein 11, THIOCYANATE ION | | Authors: | Robin, A.Y, Westphal, D, Uson, I, Czabotar, P.E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biophysical Characterization of Pro-apoptotic BimBH3 Peptides Reveals an Unexpected Capacity for Self-Association.
Structure, 29, 2021
|
|
6YRW
 
 | | SHMT from Streptococcus thermophilus Tyr55Ser variant as internal aldimine and as non-covalent complex with D-Ser | | Descriptor: | D-SERINE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2020-04-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into nucleophile substrate specificity in variants of N-Serine hydroxymethyltransferase from Streptococcus thermophilus
To Be Published
|
|
6YUG
 
 | | Crystal structure of C. parvum GNA1 in complex with acetyl-CoA and glucose 6P. | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ACETYL COENZYME *A, Diamine acetyltransferase | | Authors: | Chi, J, Cova, M, de las Rivas, M, Medina, A, Borges, R, Leivar, P, Planas, A, Uson, I, Hurtado-Guerrero, R, Izquierdo, L. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plasmodium falciparum Apicomplexan-Specific Glucosamine-6-Phosphate N -Acetyltransferase Is Key for Amino Sugar Metabolism and Asexual Blood Stage Development.
Mbio, 11, 2020
|
|
2C0H
 
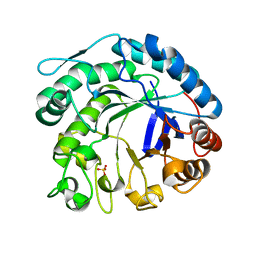 | | X-ray structure of beta-mannanase from blue mussel Mytilus edulis | | Descriptor: | MANNAN ENDO-1,4-BETA-MANNOSIDASE, SULFATE ION | | Authors: | Larsson, A.M, Anderson, L, Xu, B, Munoz, I.G, Uson, I, Janson, J.-C, Stalbrand, H, Stahlberg, J. | | Deposit date: | 2005-09-02 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-Dimensional Crystal Structure and Enzymic Characterization of Beta-Mannanase Man5A from Blue Mussel Mytilus Edulis.
J.Mol.Biol., 357, 2006
|
|
2ET0
 
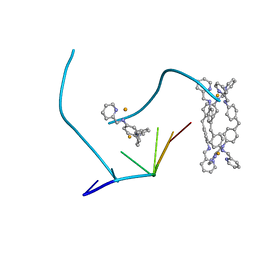 | | The structure of a three-way DNA junction in complex with a metallo-supramolecular helicate reveals a new target for drugs | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Oleksi, A, Blanco, A.G, Boer, R, Uson, I, Aymami, J, Coll, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Recognition of a Three-Way DNA Junction by a Metallosupramolecular Helicate
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
4CVD
 
 | |
4E1P
 
 | |
4E1R
 
 | |
4IJA
 
 | | Structure of S. aureus methicillin resistance factor MecR2 | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Arede, P, Botelho, T, Guevara, T, Uson, I, Oliveira, D.C, Gomis-Ruth, F.X. | | Deposit date: | 2012-12-21 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Function Studies of the Staphylococcal Methicillin Resistance Antirepressor MecR2.
J.Biol.Chem., 288, 2013
|
|
4MHX
 
 | | Crystal Structure of Sulfamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Gaertner, J, Kraetzner, R, Steinfeld, R, Sheldrick, G.M. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-14 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUN
 
 | | Structure of the N-terminal mIF4G domain from S. cerevisiae Upf2, a protein involved in the degradation of mRNAs containing premature stop codons | | Descriptor: | CHLORIDE ION, Nonsense-mediated mRNA decay protein 2 | | Authors: | Fourati, Z, Roy, B, Millan, C, Courreux, P.D, Kervestin, S, van Tilbeurgh, H, He, F, Uson, I, Jacobson, A, Graille, M. | | Deposit date: | 2013-07-25 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | A highly conserved region essential for NMD in the Upf2 N-terminal domain.
J.Mol.Biol., 426, 2014
|
|
4MIV
 
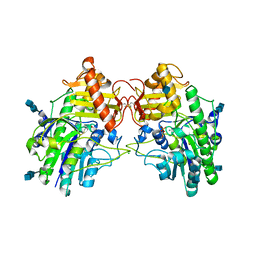 | | Crystal Structure of Sulfamidase, Crystal Form L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Sheldrick, G.M, Gaertner, J, Kraetzner, R, Steinfeld, R. | | Deposit date: | 2013-09-02 | | Release date: | 2014-05-14 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1S9B
 
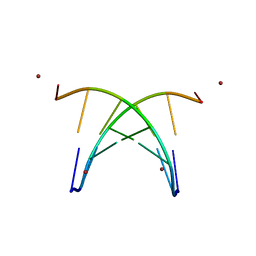 | | Crystal Structure Analysis of the B-DNA GAATTCG | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*CP*G)-3', NICKEL (II) ION | | Authors: | Valls, N, Uson, I, Gouyette, C, Subirana, J.A. | | Deposit date: | 2004-02-04 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A cubic arrangement of DNA double helices based on nickel-guanine interactions
J.Am.Chem.Soc., 126, 2004
|
|
1UNJ
 
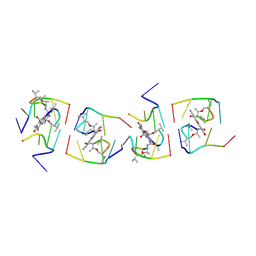 | | Crystal structure of a 7-Aminoactinomycin D complex with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINO-ACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-10 | | Release date: | 2004-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UNM
 
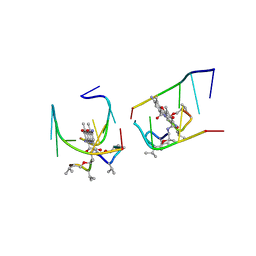 | | Crystal structure of 7-Aminoactinomycin D with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINOACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UNO
 
 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6F64
 
 | | Crystal structure of the SYCP1 C-terminal back-to-back assembly | | Descriptor: | ACETATE ION, Synaptonemal complex protein 1 | | Authors: | Dunce, J.M, Millan, C, Uson, I, Davies, O.R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-06-06 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6F63
 
 | |
