4CDG
 
 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with Nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BLOOM SYNDROME PROTEIN, NANOBODY, ... | | Authors: | Newman, J.A, Savitsky, P, Allerston, C.K, Pike, A.C.W, Pardon, E, Steyaert, J, Arrowsmith, C.H, von Delft, F, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2013-10-31 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
1KIC
 
 | | Inosine-adenosine-guanosine preferring nucleoside hydrolase from Trypanosoma vivax: Asp10Ala mutant in complex with inosine | | Descriptor: | CALCIUM ION, INOSINE, NICKEL (II) ION, ... | | Authors: | Versees, W, Decanniere, K, Van Holsbeke, E, Devroede, N, Steyaert, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzyme-substrate interactions in the purine-specific nucleoside hydrolase from Trypanosoma vivax.
J.Biol.Chem., 277, 2002
|
|
1KIE
 
 | | Inosine-adenosine-guanosine preferring nucleoside hydrolase from Trypanosoma vivax: Asp10Ala mutant in complex with 3-deaza-adenosine | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, NICKEL (II) ION, ... | | Authors: | Versees, W, Decanniere, K, Van Holsbeke, E, Devroede, N, Steyaert, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzyme-substrate interactions in the purine-specific nucleoside hydrolase from Trypanosoma vivax.
J.Biol.Chem., 277, 2002
|
|
1LOY
 
 | | X-ray structure of the H40A/E58A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
1LOW
 
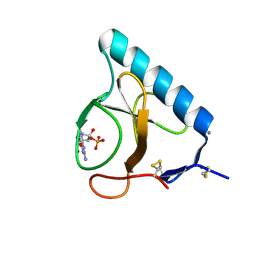 | | X-ray structure of the H40A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
1LOV
 
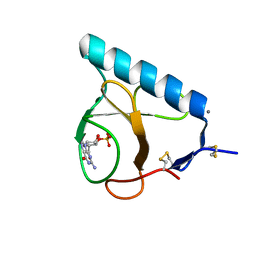 | | X-ray structure of the E58A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
6MXT
 
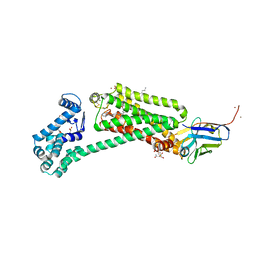 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
4GFT
 
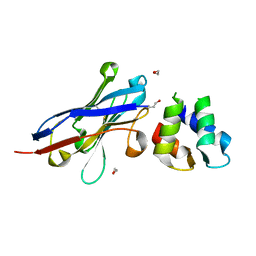 | | Malaria invasion machinery protein-Nanobody complex | | Descriptor: | 1,2-ETHANEDIOL, Myosin A tail domain interacting protein, Nanobody | | Authors: | Khamrui, S, Turley, S, Pardon, E, Steyaert, J, Verlinde, C, Fan, E, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-03 | | Release date: | 2013-07-03 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
4I1N
 
 | |
4I13
 
 | |
7OCJ
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
4KML
 
 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the full-length human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-05-08 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
4KSD
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A, R2 protein | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (4.1001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSC
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSB
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KDT
 
 | | Structure of an early native-like intermediate of beta2-microglobulin amyloidosis | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Nanobody24, ... | | Authors: | Vanderhaegen, S, Fislage, M, Pardon, E, Versees, W, Steyaert, J. | | Deposit date: | 2013-04-25 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an early native-like intermediate of beta 2-microglobulin amyloidogenesis.
Protein Sci., 22, 2013
|
|
8T60
 
 | |
6VBG
 
 | | Lactose permease complex with thiodigalactoside and nanobody 9043 | | Descriptor: | Galactoside permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose, nanobody 9043, ... | | Authors: | Kumar, H, Stroud, R.M, Kaback, H.R, Finer-Moore, J, Smirnova, I, Kasho, V, Pardon, E, Steyart, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diversity in kinetics correlated with structure in nano body-stabilized LacY.
Plos One, 15, 2020
|
|
7SP8
 
 | | Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SPA
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP9
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP7
 
 | | Chlorella virus hyaluronan synthase inhibited by UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP6
 
 | | Chlorella virus hyaluronan synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
6NFJ
 
 | |
6TYL
 
 | | Crystal structure of mammalian Ric-8A:Galpha(i):nanobody complex | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Nanobody A, Nanobody B, ... | | Authors: | Mou, T.C, McClelland, L, Yates-Hansen, C, Sprang, S.R. | | Deposit date: | 2019-08-09 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
