1JN4
 
 | | The Crystal Structure of Ribonuclease A in complex with 2'-deoxyuridine 3'-pyrophosphate (P'-5') adenosine | | Descriptor: | ADENOSINE-5'-[TRIHYDROGEN DIPHOSPHATE] P'-3'-ESTER WITH 2'-DEOXYURIDINE, Pancreatic Ribonuclease A | | Authors: | Jardine, A.M, Leonidas, D.D, Jenkins, J.L, Park, C, Raines, R.T, Acharya, K.R, Shapiro, R. | | Deposit date: | 2001-07-23 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cleavage of 3',5'-Pyrophosphate-Linked Dinucleotides by Ribonuclease A and Angiogenin
Biochemistry, 40, 2001
|
|
1R5H
 
 | | Crystal Structure of MetAP2 complexed with A320282 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R58
 
 | | Crystal Structure of MetAP2 complexed with A357300 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-((2S,3R)-3-AMINO-2-HYDROXY-5-(ISOPROPYLSULFANYL)PENTANOYL)-N-3-CHLOROBENZOYL HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Ericken, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R5G
 
 | | Crystal Structure of MetAP2 complexed with A311263 | | Descriptor: | (2S,3R)-3-AMINO-2-HYDROXY-5-(ETHYLSULFANYL)PENTANOYL-((S)-(-)-(1-NAPHTHYL)ETHYL)AMIDE, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
7C0J
 
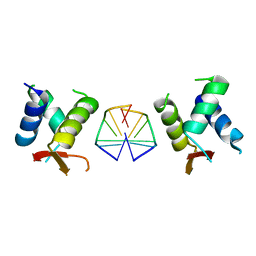 | | Crystal structure of chimeric mutant of GH5 in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Histone H5,Double-stranded RNA-specific adenosine deaminase | | Authors: | Choi, H.J, Park, C.H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
7C0I
 
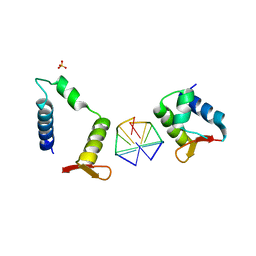 | | Crystal structure of chimeric mutant of E3L in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-binding protein,Double-stranded RNA-specific adenosine deaminase, SULFATE ION | | Authors: | Choi, H.J, Park, C.H, Kim, J.S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
8USQ
 
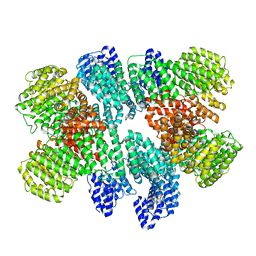 | |
8USP
 
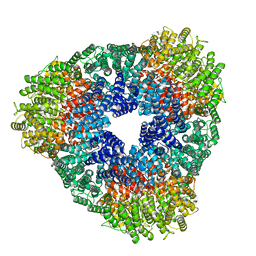 | |
6CEZ
 
 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V2 Fab 16C2 in complex with V2 peptide ConB | | Descriptor: | HIV-1 gp120 V2 Peptide Con B, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2, Light chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2 | | Authors: | Kong, X, Pan, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Select gp120 V2 domain specific antibodies derived from HIV and SIV infection and vaccination inhibit gp120 binding to alpha 4 beta 7.
PLoS Pathog., 14, 2018
|
|
3NR1
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | Descriptor: | HD domain-containing protein 3, MANGANESE (II) ION | | Authors: | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
2RVC
 
 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | Descriptor: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | Authors: | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | Deposit date: | 2015-07-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
2A4F
 
 | | Synthesis and Activity of N-Axyl Azacyclic Urea HIV-1 Protease Inhibitors with High Potency Against Multiple Drug Resistant Viral Strains. | | Descriptor: | (5R,6R)-5-BENZYL-6-HYDROXY-2,4-BIS(4-HYDROXY-3-METHOXYBENZYL)-1-[3-(4-HYDROXYPHENYL)PROPANOYL]-1,2,4-TRIAZEPAN-3-ONE, Pol polyprotein | | Authors: | Zhao, C, Sham, H, Sun, M, Lin, S, Stoll, V, Stewart, K.D, Mo, H, Vasavanonda, S, Saldivar, A, McDonald, E. | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and activity of N-acyl azacyclic urea HIV-1 protease inhibitors with high potency against multiple drug resistant viral strains
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3NQW
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | Descriptor: | CG11900, MANGANESE (II) ION, SULFATE ION | | Authors: | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
2QF6
 
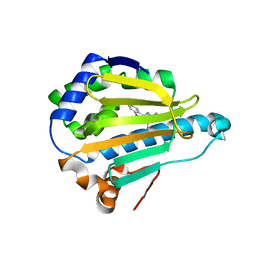 | | HSP90 complexed with A56322 | | Descriptor: | 6-(3-BROMO-2-NAPHTHYL)-1,3,5-TRIAZINE-2,4-DIAMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG2
 
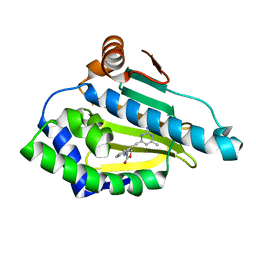 | | HSP90 complexed with A917985 | | Descriptor: | 3-({2-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)ETHYNYL]BENZYL}AMINO)-1,3-OXAZOL-2(3H)-ONE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG0
 
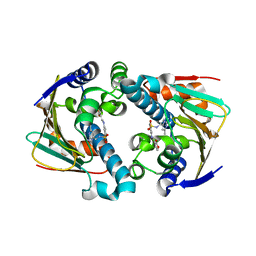 | | HSP90 complexed with A943037 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)METHYL]-3-{[(E)-(2-OXODIHYDROFURAN-3(2H)-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QFO
 
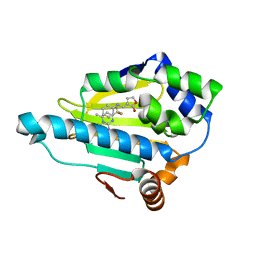 | | HSP90 complexed with A143571 and A516383 | | Descriptor: | (3E)-3-[(phenylamino)methylidene]dihydrofuran-2(3H)-one, 4-METHYL-6-(TRIFLUOROMETHYL)PYRIMIDIN-2-AMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
CHEM.BIOL.DRUG DES., 70, 2007
|
|
1RPJ
 
 | | CRYSTAL STRUCTURE OF D-ALLOSE BINDING PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (PRECURSOR OF PERIPLASMIC SUGAR RECEPTOR), SULFATE ION, ZINC ION, ... | | Authors: | Chaudhuri, B, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1999-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of D-allose binding protein from Escherichia coli bound to D-allose at 1.8 A resolution.
J.Mol.Biol., 286, 1999
|
|
5VKC
 
 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
2EA2
 
 | | h-MetAP2 complexed with A773812 | | Descriptor: | 3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}-2-METHYLBENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EA4
 
 | | h-MetAP2 complexed with A797859 | | Descriptor: | 2-(2-AMINOETHOXY)-3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7W3D
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Descriptor: | Bromodomain-containing protein 4, N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of BET specific bromodomain inhibitors with a novel scaffold.
Bioorg.Med.Chem., 72, 2022
|
|
1EA9
 
 | | Cyclomaltodextrinase | | Descriptor: | CYCLOMALTODEXTRINASE | | Authors: | Cho, H.-S, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cyclomaltodextrinase, Neopullulanase, and Maltogenic Amylase are Nearly Indistinguishable from Each Other
J.Biol.Chem., 277, 2002
|
|
3KEN
 
 | | Human Eg5 in complex with S-trityl-L-cysteine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Parke, C.L, Wojcik, E.J, Kim, S, Worthylake, D.K. | | Deposit date: | 2009-10-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core.
J.Biol.Chem., 285, 2010
|
|
3L3L
 
 | | PARP complexed with A906894 | | Descriptor: | 3-oxo-2-piperidin-4-yl-2,3-dihydro-1H-isoindole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
