1BGI
 
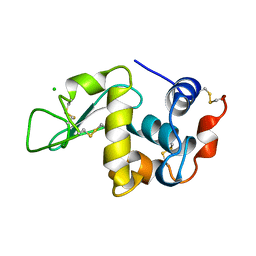 | | ORTHORHOMBIC LYSOZYME CRYSTALLIZED AT HIGH TEMPERATURE (310K) | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Oki, H, Matsuura, Y, Komatsu, H, Chernov, A.A. | | Deposit date: | 1998-05-28 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structure of orthorhombic lysozyme crystallized at high temperature: correlation between morphology and intermolecular contacts.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7W64
 
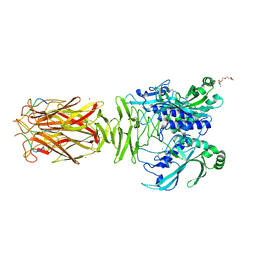 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with N-terminal peptide fragment of TcpF | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W65
 
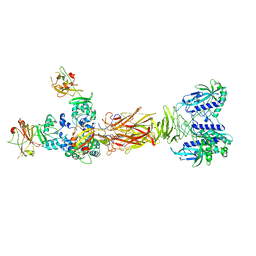 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with secreted protein TcpF | | Descriptor: | Toxin coregulated pilus biosynthesis protein F, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W63
 
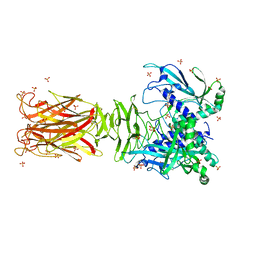 | | Crystal structure of minor pilin TcpB from Vibrio cholerae | | Descriptor: | SULFATE ION, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
5YQ0
 
 | | Crystal structure of secreted protein CofJ from ETEC. | | Descriptor: | CALCIUM ION, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YPZ
 
 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | Descriptor: | CofB, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2EV8
 
 | |
1A13
 
 | | G PROTEIN-BOUND CONFORMATION OF MASTOPARAN-X, NMR, 14 STRUCTURES | | Descriptor: | MASTOPARAN-X | | Authors: | Kusunoki, H, Wakamatsu, K, Sato, K, Miyazawa, T, Kohno, T. | | Deposit date: | 1997-12-20 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | G protein-bound conformation of mastoparan-X: heteronuclear multidimensional transferred nuclear overhauser effect analysis of peptide uniformly enriched with 13C and 15N.
Biochemistry, 37, 1998
|
|
2RQ1
 
 | |
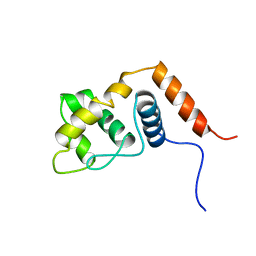
2RQ5
 
 | |
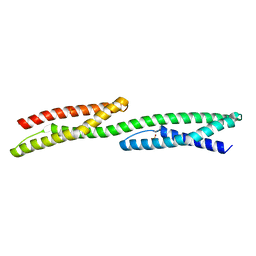
1S35
 
 | |
1U4Q
 
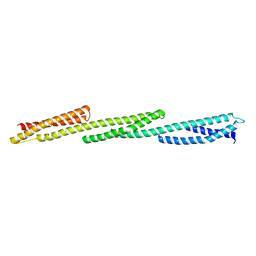 | | Crystal Structure of Repeats 15, 16 and 17 of Chicken Brain Alpha Spectrin | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-26 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
1U5P
 
 | | Crystal Structure of Repeats 15 and 16 of Chicken Brain Alpha Spectrin | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Spectrin alpha chain, ... | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
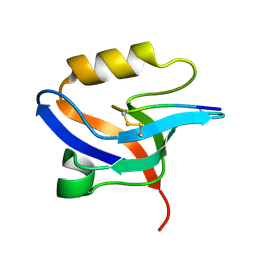
2EJY
 
 | |
1K1V
 
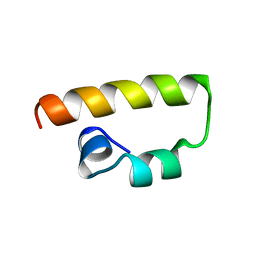 | | Solution Structure of the DNA-Binding Domain of MafG | | Descriptor: | MafG | | Authors: | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
7XW8
 
 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with TAK-418 distomer, FAD-adduct | | Descriptor: | GLYCEROL, Lysine-specific histone demethylase 1A, MAGNESIUM ION, ... | | Authors: | Oki, H. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design, synthesis, and structure-activity relationship of TAK-418 and its derivatives as a novel series of LSD1 inhibitors with lowered risk of hematological side effects.
Eur.J.Med.Chem., 239, 2022
|
|
5B5O
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-phenyl-4-((4H-1,2,4-triazol-3-ylsulfanyl)methyl)-1,3-thiazol-2-amine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
5B5P
 
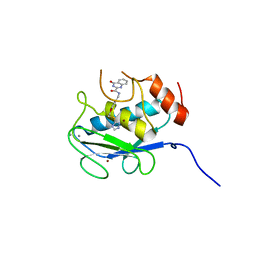 | | Crystal structure of the catalytic domain of MMP-13 complexed with 4-oxo-N-(3-(2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy)benzyl)-3,4-dihydroquinazoline-2-carboxamide | | Descriptor: | 4-oxo-N-{3-[2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy]benzyl}-3,4-dihydroquinazoline-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
5AXQ
 
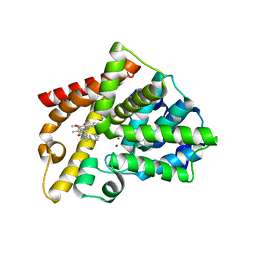 | | Crystal structure of the catalytic domain of PDE10A complexed with highly potent and brain-penetrant PDE10A Inhibitor with 2-oxindole scaffold | | Descriptor: | 1-(cyclopropylmethyl)-4-fluoranyl-5-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]-3,3-dimethyl-indol-2-one, 3-[3-fluoranyl-4-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]phenyl]-1,3-oxazolidin-2-one, MAGNESIUM ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design and synthesis of a novel 2-oxindole scaffold as a highly potent and brain-penetrant phosphodiesterase 10A inhibitor
Bioorg.Med.Chem., 23, 2015
|
|
5AXP
 
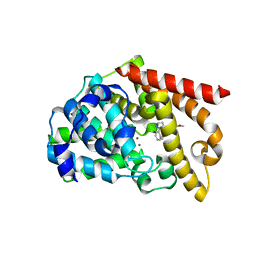 | | Crystal structure of the catalytic domain of PDE10A complexed with 1-(2-fluoro-4-(2-oxo-1,3-oxazolidin-3-yl)phenyl)-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 3-[3-fluoranyl-4-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]phenyl]-1,3-oxazolidin-2-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and synthesis of a novel 2-oxindole scaffold as a highly potent and brain-penetrant phosphodiesterase 10A inhibitor
Bioorg.Med.Chem., 23, 2015
|
|
5B4O
 
 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
7E0G
 
 | |
5B4L
 
 | | Crystal structure of the catalytic domain of human PDE10A complexed with 1-(cyclopropylmethyl)-5-(2-(2,3-dihydro-1H-imidazo[1,2-a]benzimidazol-1-yl)ethoxy)-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 1-(cyclopropylmethyl)-5-(2-(2,3-dihydro-1H-imidazo[1,2-a]benzimidazol-1-yl)ethoxy)-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent and selective pyridazin-4(1H)-one-based PDE10A inhibitors interacting with Tyr683 in the PDE10A selectivity pocket
Bioorg.Med.Chem., 24, 2016
|
|
5B4K
 
 | | Crystal structure of the catalytic domain of human PDE10A complexed with N-(4-((5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy)phenyl)-1H-benzimidazol-2-amine | | Descriptor: | MAGNESIUM ION, N-(4-((5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy)phenyl)-1H-benzimidazol-2-amine, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and synthesis of potent and selective pyridazin-4(1H)-one-based PDE10A inhibitors interacting with Tyr683 in the PDE10A selectivity pocket
Bioorg.Med.Chem., 24, 2016
|
|
5XKM
 
 | | Crystal structure of human phosphodiesterase 2A in complex with 6-methyl-N-(1-(4-(trifluoromethoxy)phenyl)propyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | 6-methyl-N-[(1R)-1-[4-(trifluoromethyloxy)phenyl]propyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Kondo, M, Snell, G, Lane, W. | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of an Orally Bioavailable, Brain-Penetrating, in Vivo Active Phosphodiesterase 2A Inhibitor Lead Series for the Treatment of Cognitive Disorders.
J. Med. Chem., 60, 2017
|
|
