1I5N
 
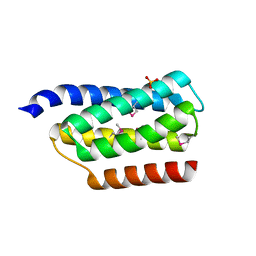 | | Crystal structure of the P1 domain of CheA from Salmonella typhimurium | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, SULFATE ION | | Authors: | Mourey, L, Da Re, S, Pedelacq, J.-D, Tolstyk, T, Faurie, C, Guillet, V, Stock, J.B, Samama, J.-P. | | Deposit date: | 2001-02-28 | | Release date: | 2001-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis
J.Biol.Chem., 276, 2001
|
|
1ATT
 
 | |
1AVB
 
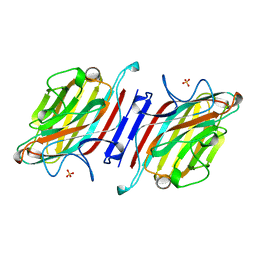 | | ARCELIN-1 FROM PHASEOLUS VULGARIS L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARCELIN-1, ... | | Authors: | Mourey, L, Pedelacq, J.D, Fabre, C, Rouge, P, Samama, J.P. | | Deposit date: | 1997-09-15 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the arcelin-1 dimer from Phaseolus vulgaris at 1.9-A resolution.
J.Biol.Chem., 273, 1998
|
|
1BUL
 
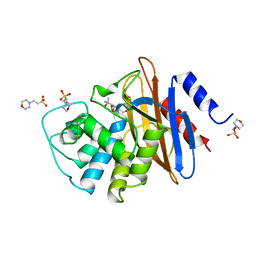 | | 6ALPHA-(HYDROXYPROPYL)PENICILLANATE ACYLATED ON NMC-A BETA-LACTAMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | 2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NMC-A BETA-LACTAMASE | | Authors: | Mourey, L, Swaren, P, Miyashita, K, Bulychev, A, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-04 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Inhibition of the Nmc-A B-Lactamase by a Penicillanic Acid Derivative, and the Structural Bases for the Increase in Substrate Profile of This Antibiotic Resistance Enzyme
J.Am.Chem.Soc., 120, 1998
|
|
2QK7
 
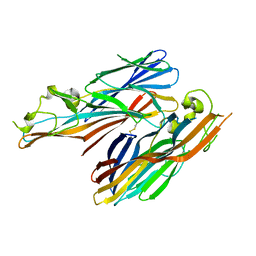 | | A covalent S-F heterodimer of staphylococcal gamma-hemolysin | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Roblin, P, Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2007-07-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of beta-barrel pore-forming toxins.
Proteins, 71, 2008
|
|
1T5R
 
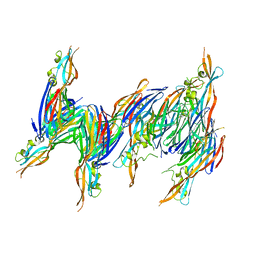 | | STRUCTURE OF THE PANTON-VALENTINE LEUCOCIDIN S COMPONENT FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | LukS-PV | | Authors: | Guillet, V, Roblin, P, Keller, D, Prevost, G, Mourey, L. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of leucotoxin S component: new insight into the Staphylococcal beta-barrel pore-forming toxins.
J.Biol.Chem., 279, 2004
|
|
6RCX
 
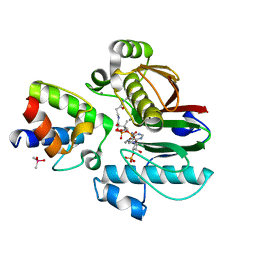 | | Mycobacterial 4'-phosphopantetheinyl transferase PptAb in complex with the ACP domain of PpsC. | | Descriptor: | CACODYLATE ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
4IYC
 
 | | Structure of the T244A mutant of the PANTON-VALENTINE LEUCOCIDIN component from STAPHYLOCOCCUS AUREUS | | Descriptor: | LukS-PV | | Authors: | Maveyraud, L, Guerin, F, Lavnetie, B.J, Prevost, G, Mourey, L. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
3CA4
 
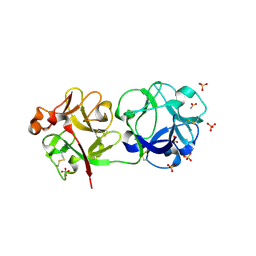 | | Sambucus nigra agglutinin II, tetragonal crystal form- complexed to lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA0
 
 | | Sambucus nigra agglutinin II (SNA-II), hexagonal crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Agglutinin II, ... | | Authors: | Maveyraud, L, Niwa, H, Guillet, V, Palmer, R.A, Reynolds, C.D, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA5
 
 | | Crystal structure of Sambucus nigra agglutinin II (SNA-II)-tetragonal crystal form- complexed to alpha1 methylgalactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CAH
 
 | | Sambucus nigra aggutinin II. tetragonal crystal form- complexed to fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-20 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA1
 
 | | Sambucus nigra agglutinin II (SNA-II)- tetragonal crystal form- complexed to galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3C9Z
 
 | | Sambucus nigra agglutinin II (SNA-II), tetragonal crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Guillet, V, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
5L84
 
 | |
1PVL
 
 | | STRUCTURE OF THE PANTON-VALENTINE LEUCOCIDIN F COMPONENT FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LEUCOCIDIN | | Authors: | Pedelacq, J.D, Mourey, L, Maveyraud, L, Prevost, G, Samama, J.P. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a Staphylococcus aureus leucocidin component (LukF-PV) reveals the fold of the water-soluble species of a family of transmembrane pore-forming toxins.
Structure Fold.Des., 7, 1999
|
|
4FLQ
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290I from Neisseria polysaccharea. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLO
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
6SCR
 
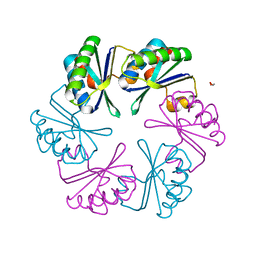 | |
4N2Z
 
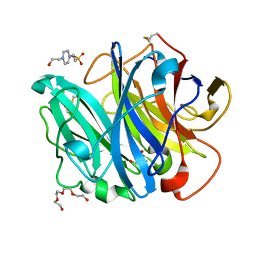 | | Crystal Structure of the alpha-L-arabinofuranosidase PaAbf62A from Podospora anserina in complex with cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
4N1I
 
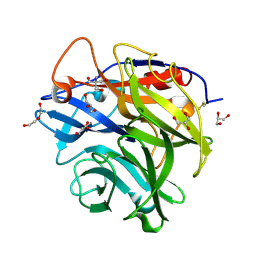 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maidys | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-15 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
4N2R
 
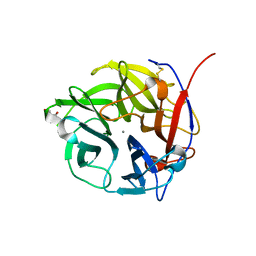 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maydis in complex with L-arabinofuranose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, alpha-L-arabinofuranose, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
4J0O
 
 | |
4IYT
 
 | | Structure Of The Y184A Mutant Of The PANTON-VALENTINE LEUCOCIDIN S Component From STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LukS-PV | | Authors: | Guerin, F, Laventie, B.J, Prevost, G, Mourey, L, Maveyraud, L. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
4IYA
 
 | | Structure of the Y250A mutant of the PANTON-VALENTINE LEUCOCIDIN S component from STAPHYLOCOCCUS AUREUS | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, LukS-PV | | Authors: | Maveyraud, L, Guerin, F, Laventie, B.J, Prevost, G, Mourey, L. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
