2J0E
 
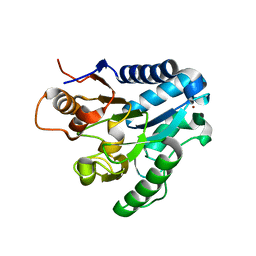 | | Three dimensional structure and catalytic mechanism of 6- phosphogluconolactonase from Trypanosoma brucei | | Descriptor: | 6-PHOSPHOGLUCONOLACTONASE, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Delarue, M, Duclert-Savatier, N, Miclet, E, Haouz, A, Giganti, D, Ouazzani, J, Lopez, P, Nilges, M, Stoven, V. | | Deposit date: | 2006-08-02 | | Release date: | 2007-01-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Dimensional Structure and Implications for the Catalytic Mechanism of 6-Phosphogluconolactonase from Trypanosoma Brucei.
J.Mol.Biol., 366, 2007
|
|
6V4B
 
 | | DeCLIC N-terminal Domain 34-202 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Neur_chan_LBD domain-containing protein, ... | | Authors: | Delarue, M, Hu, H.D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for allosteric transitions of a multidomain pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V4A
 
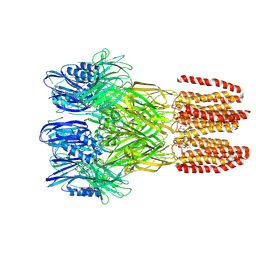 | |
6V4S
 
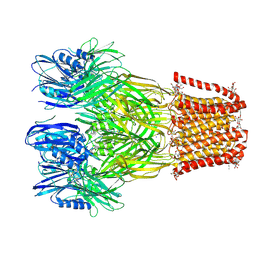 | |
1JMS
 
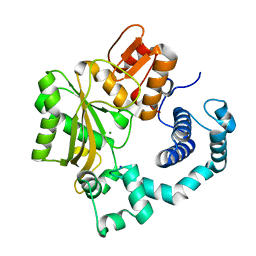 | | Crystal Structure of the Catalytic Core of Murine Terminal Deoxynucleotidyl Transferase | | Descriptor: | MAGNESIUM ION, SODIUM ION, TERMINAL DEOXYNUCLEOTIDYLTRANSFERASE | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Sukumar, N, Jourdan, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-07-19 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
Embo J., 21, 2002
|
|
1KDH
 
 | | Binary Complex of Murine Terminal Deoxynucleotidyl Transferase with a Primer Single Stranded DNA | | Descriptor: | 5'-D(P*(BRU)P*(BRU)P*(BRU)P*(BRU))-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
1KEJ
 
 | | Crystal Structure of Murine Terminal Deoxynucleotidyl Transferase complexed with ddATP | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
1ATT
 
 | |
7PBK
 
 | | Vibriophage phiVC8 family A DNA polymerase (DpoZ), two conformations: thumb-exo open and thumb-exo closed | | Descriptor: | DNA polymerase I | | Authors: | Czernecki, D, Hu, H, Romoli, F, Delarue, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage phi VC8.
Nucleic Acids Res., 49, 2021
|
|
1G3U
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) | | Descriptor: | MAGNESIUM ION, SULFATE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Li de la Sierra, I, Munier-Lehmann, H, Gilles, A.M, Barzu, O, Delarue, M. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray structure of TMP kinase from Mycobacterium tuberculosis complexed with TMP at 1.95 A resolution.
J.Mol.Biol., 311, 2001
|
|
1GSI
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) | | Descriptor: | ACETATE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ursby, T, Weik, M, Fioravanti, E, Delarue, M, Goeldner, M, Bourgeois, D. | | Deposit date: | 2002-01-03 | | Release date: | 2002-03-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cryophotolysis of Caged Compounds: A Technique for Trapping Intermediate States in Protein Crystals
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GTV
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE-5'-DIPHOSPHATE (TDP) | | Descriptor: | ACETATE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ursby, T, Weik, M, Fioravanti, E, Delarue, M, Goeldner, M, Bourgeois, D. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-28 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cryophotolysis of Caged Compounds: A Technique for Trapping Intermediate States in Protein Crystals
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4X5T
 
 | | alpha 1 glycine receptor transmembrane structure fused to the extracellular domain of GLIC | | Descriptor: | ACETATE ION, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Sauguet, L, Corringer, P.J, Huon, C, Delarue, M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Allosteric and hyperekplexic mutant phenotypes investigated on an alpha 1 glycine receptor transmembrane structure.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7ODY
 
 | |
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7X
 
 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
1N5J
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE DIPHOSPHATE (TDP) AND THYMIDINE TRIPHOSPHATE (TTP) AT PH 5.4 (1.85 A RESOLUTION) | | Descriptor: | MAGNESIUM ION, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 327, 2003
|
|
1N5I
 
 | | CRYSTAL STRUCTURE OF INACTIVE MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) AT PH 4.6 (RESOLUTION 1.85 A) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRATE ANION, SULFATE ION, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the Reaction Pathway
J.Mol.Biol., 327, 2003
|
|
1N5K
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE CRYSTALLIZED IN SODIUM MALONATE (RESOLUTION 2.1 A) | | Descriptor: | ACETATE ION, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 375, 2003
|
|
1N5L
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE CRYSTALLIZED IN SODIUM MALONATE, AFTER CATALYSIS IN THE CRYSTAL (2.3 A RESOLUTION) | | Descriptor: | ACETATE ION, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 375, 2003
|
|
6T7Y
 
 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
1EQR
 
 | | CRYSTAL STRUCTURE OF FREE ASPARTYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTYL-TRNA SYNTHETASE, MAGNESIUM ION | | Authors: | Rees, B, Webster, G, Delarue, M, Boeglin, M, Moras, D. | | Deposit date: | 2000-04-06 | | Release date: | 2000-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aspartyl tRNA-synthetase from Escherichia coli: flexibility and adaptability to the substrates.
J.Mol.Biol., 299, 2000
|
|
6ZPA
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and one catalytic Zn2+ ion | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (0.86000258 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPC
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.2683593 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPB
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and two catalytic Co2+ ions | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, COBALT (II) ION, DatZ | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.72097385 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
