2M8V
 
 | | Solution Structure and Activity Study of Bovicin HJ50, a Particular Type AII Lantibiotic | | Descriptor: | BovA | | Authors: | Zhang, J, Feng, Y, Wang, J, Zhong, J. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Type AII lantibiotic bovicin HJ50 with a rare disulfide bond: structure, structure-activity relationships and mode of action.
Biochem.J., 461, 2014
|
|
2MJV
 
 | |
5H14
 
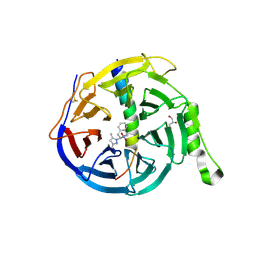 | | EED in complex with an allosteric PRC2 inhibitor EED666 | | Descriptor: | 2-[3-(3,5-dimethylpyrazol-1-yl)-4-nitro-phenyl]-3,4-dihydro-1H-isoquinoline, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H15
 
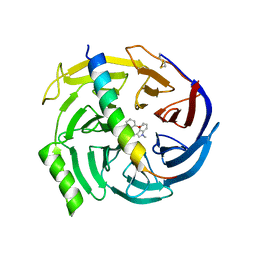 | | EED in complex with PRC2 allosteric inhibitor EED709 | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
5H13
 
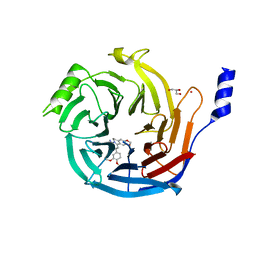 | | EED in complex with PRC2 allosteric inhibitor EED396 | | Descriptor: | 4-azanylidene-2-(3-methoxy-4-propan-2-yloxy-phenyl)-6,7-dihydro-[1,3]benzodioxolo[6,5-a]quinolizine-3-carbonitrile, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H17
 
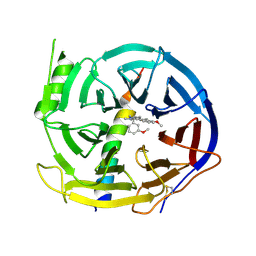 | | EED in complex with PRC2 allosteric inhibitor EED210 | | Descriptor: | (3R,4aS,10aS)-6-methoxy-3-[(3-methoxyphenyl)methyl]-1-methyl-3,4,4a,5,10,10a-hexahydro-2H-benzo[g]quinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
2F90
 
 | | Crystal structure of bisphosphoglycerate mutase in complex with 3-phosphoglycerate and AlF4- | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, TETRAFLUOROALUMINATE ION | | Authors: | Wang, Y, Liu, L, Wei, Z, Gong, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase
J.Biol.Chem., 281, 2006
|
|
2H4Z
 
 | |
2H4X
 
 | |
2HHJ
 
 | | Human bisphosphoglycerate mutase complexed with 2,3-bisphosphoglycerate (15 days) | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, ... | | Authors: | Wang, Y, Gong, W. | | Deposit date: | 2006-06-28 | | Release date: | 2006-10-24 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
J.Biol.Chem., 281, 2006
|
|
3NDP
 
 | | Crystal structure of human AK4(L171P) | | Descriptor: | Adenylate kinase isoenzyme 4, SULFATE ION | | Authors: | Liu, R, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human adenylate kinase 4 (L171P) suggests the role of hinge region in protein domain motion
Biochem.Biophys.Res.Commun., 379, 2009
|
|
2HNK
 
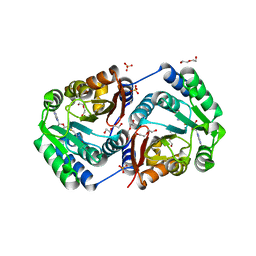 | | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent O-methyltransferase, ... | | Authors: | Hou, X, Wei, Z, Gong, W. | | Deposit date: | 2006-07-13 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans.
J.Struct.Biol., 159, 2007
|
|
2I6K
 
 | | Crystal structure of human type I IPP isomerase complexed with a substrate analog | | Descriptor: | ACETIC ACID, AMINOETHANOLPYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase 1, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
2WLU
 
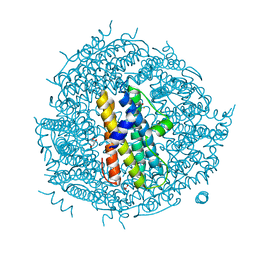 | | Iron-bound crystal structure of Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-15 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
2WLA
 
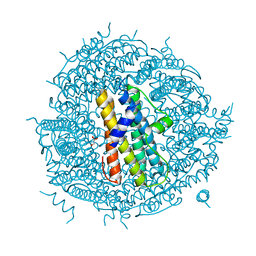 | | Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, GLYCEROL | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-23 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
5FEJ
 
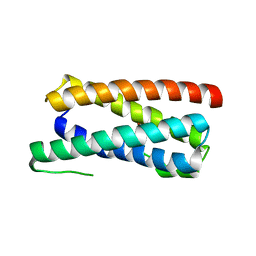 | | CopM in the Cu(I)-bound form | | Descriptor: | COPPER (I) ION, CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-17 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFA
 
 | |
5FFC
 
 | | CopM in the Cu(II)-bound form | | Descriptor: | COPPER (II) ION, CopM, SULFATE ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFE
 
 | | CopM in the Ag-bound form (by soaking) | | Descriptor: | CopM, SILVER ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFB
 
 | | CopM in the apo form | | Descriptor: | CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
