2J3J
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
5YK6
 
 | | Crystal Structure of Mmm1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Maintenance of mitochondrial morphology protein 1 | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YK7
 
 | | Crystal Structure of Mdm12-Mmm1 complex | | Descriptor: | Maintenance of mitochondrial morphology protein 1, Mitochondrial distribution and morphology protein 12, PHOSPHATE ION | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.799 Å) | | Cite: | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4MLG
 
 | | Structure of RS223-Beta-xylosidase | | Descriptor: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | Authors: | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | Deposit date: | 2013-09-06 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of RS223-Beta-xylosidase
To be Published
|
|
7EXP
 
 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | Descriptor: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
2KCC
 
 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
6L2U
 
 | | Soluble methane monooxygenase reductase FAD-binding domain from Methylosinus sporium. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methane monooxygenase | | Authors: | Park, J.H, Ha, S.C, Rao, Z, Yoo, H, Yoon, C, Kim, S.Y, Kim, D.S, Lee, S.J. | | Deposit date: | 2019-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5.
Dalton Trans, 50, 2021
|
|
6X84
 
 | |
3L9P
 
 | |
7BWL
 
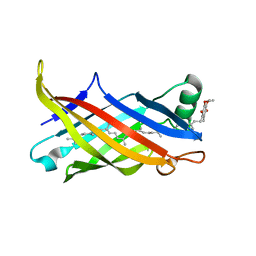 | | Structure of antibiotic sequester from Pseudomonas aerurinosa | | Descriptor: | UPF0312 protein PA0423, Ubiquinone-8 | | Authors: | Hong, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Pseudomonas aeruginosa PA0423 protein and its functional implication in antibiotic sequestration.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
2LKP
 
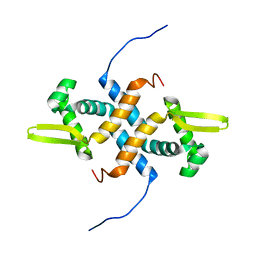 | | solution structure of apo-NmtR | | Descriptor: | HTH-type transcriptional regulator NmtR | | Authors: | Lee, C, Giedroc, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mycobacterium tuberculosis NmtR in the Apo State: Insights into Ni(II)-Mediated Allostery.
Biochemistry, 51, 2012
|
|
5ZQH
 
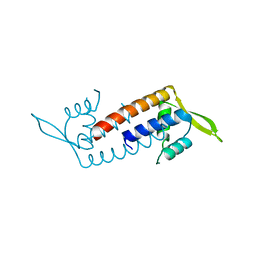 | | Crystal structure of Streptococcus transcriptional regulator | | Descriptor: | PadR family transcriptional regulator | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based functional analysis of a PadR transcription factor from Streptococcus pneumoniae and characteristic features in the PadR subfamily-2.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
2L14
 
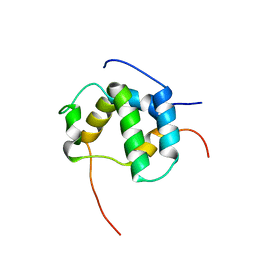 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
2L6I
 
 | |
8SD7
 
 | | Carbonic anhydrase II radiation damage RT 61-90 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SF1
 
 | | Carbonic anhydrase II XFEL radiation damage RT | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD9
 
 | | Carbonic anhydrase II radiation damage RT 121-150 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD8
 
 | | Carbonic anhydrase II radiation damage RT 91-120 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD6
 
 | | Carbonic anhydrase II radiation damage RT 31-60 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD1
 
 | | Carbonic anhydrase II radiation damage RT 1-30 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.C, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
1V3A
 
 | |
7RUN
 
 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrrolo[2,3-d]pyrimidine inhibitor. | | Descriptor: | 1-(4-{(1s,3s)-3-[4-amino-5-(3-amino-4-chlorophenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl]cyclobutyl}piperazin-1-yl)ethan-1-one, CHLORIDE ION, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2021-08-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Antitarget Selectivity and Tolerability of Novel Pyrrolo[2,3- d ]pyrimidine RET Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6VHG
 
 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 3-(3,4-dimethoxyphenyl)-N~5~-(1-methylpiperidin-4-yl)-6-phenylpyrazolo[1,5-a]pyrimidine-5,7-diamine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Efficacy and Tolerability of Pyrazolo[1,5-a]pyrimidine RET Kinase Inhibitors for the Treatment of Lung Adenocarcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
5GPG
 
 | | Co-crystal structure of the FK506 binding domain of human FKBP25, Rapamycin and the FRB domain of human mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP3, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Lee, H.B, Lee, S.Y, Rhee, H.W, Lee, C.W. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Acs Cent.Sci., 2, 2016
|
|
