4Z86
 
 | |
3R00
 
 | |
5B6J
 
 | |
3R01
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-bromo-7-methoxy-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Liu, J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3VBX
 
 | |
3VC4
 
 | |
3VBT
 
 | |
3VBV
 
 | |
3VBQ
 
 | |
3VBY
 
 | |
3VBW
 
 | |
7USZ
 
 | | Human DDAH-1, holo (Zn-bound) form | | Descriptor: | CHLORIDE ION, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, ZINC ION | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7UT0
 
 | | Human DDAH-1, apo form | | Descriptor: | 1,2-ETHANEDIOL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
5JBQ
 
 | |
5YOZ
 
 | |
5ZK0
 
 | | Crystal structure of Peptidyl-tRNA hydrolase mutant -M71A from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Shahid, S, Kabra, A, Pal, R.K, Arora, A. | | Deposit date: | 2018-03-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Role of methionine 71 in substrate recognition and structural integrity of bacterial peptidyl-tRNA hydrolase.
Biochim. Biophys. Acta, 1866, 2018
|
|
5VF5
 
 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5YNR
 
 | |
2MP4
 
 | | Solution Structure of ADF like UNC-60A Protein of Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 1, isoforms a/b | | Authors: | Shukla, V, Yadav, R, Kabra, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2014-05-11 | | Release date: | 2014-06-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Backbone dynamics of ADF like UNC-60A protein from Caenorhabditis elegans: its divergence from conventional ADF/cofilin
To be Published
|
|
3OYO
 
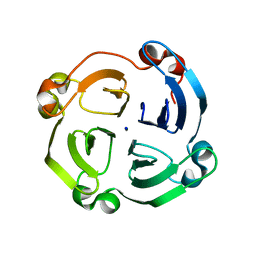 | | Crystal structure of hemopexin fold protein CP4 from cow pea | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gaur, V, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-09-23 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a haemopexin-fold protein from cow pea (Vigna unguiculata) suggests functional diversity of haemopexins in plants
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3U6B
 
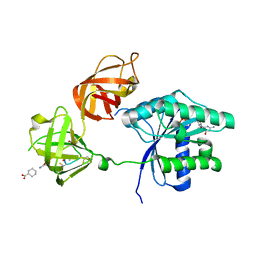 | | Ef-tu (escherichia coli) in complex with nvp-ldi028 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
2MV2
 
 | |
6ISJ
 
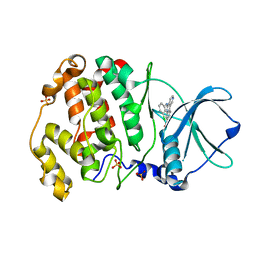 | | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Cho, H.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans
To be published
|
|
7JHD
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with TTC-352 and GRIP Peptide | | Descriptor: | 3-(4-fluorophenyl)-2-(4-hydroxyphenoxy)-1-benzothiophene-6-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Fanning, S.W, Abderraman, B, Maximov, P.Y, Jordan, V.C, Greene, G.L. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Rapid Induction of the Unfolded Protein Response and Apoptosis by Estrogen Mimic TTC-352 for the Treatment of Endocrine-Resistant Breast Cancer.
Mol.Cancer Ther., 20, 2021
|
|
7JKB
 
 | | 2xVH Fab | | Descriptor: | Anti-Her2, Anti-lysozyme | | Authors: | Lord, D.M, Zhou, Y.F. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Bringing the Heavy Chain to Light: Creating a Symmetric, Bivalent IgG-Like Bispecific.
Antibodies, 9, 2020
|
|
