8F60
 
 | | anti-BTLA monoclonal antibody r23C8 in complex with BTLA | | Descriptor: | ACETATE ION, B- and T-lymphocyte attenuator, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6O
 
 | | anti-BTLA monoclonal antibody h22B3 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h22B3 Fab heavy chain, h22B3 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6L
 
 | | anti-BTLA monoclonal antibody h25F7 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h25F7 Fab heavy chain, h25F7 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
5LOL
 
 | | Glutathione-bound Dehydroascorbate Reductase 2 of Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase DHAR2, ... | | Authors: | Young, D.R, Pallo, A, Bodra, N, Messens, J. | | Deposit date: | 2016-08-09 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arabidopsis thaliana dehydroascorbate reductase 2: Conformational flexibility during catalysis.
Sci Rep, 7, 2017
|
|
8FHS
 
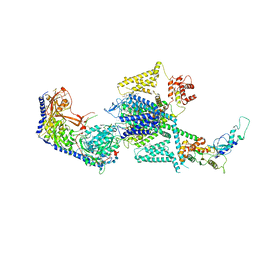 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of amiodarone and sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8GXM
 
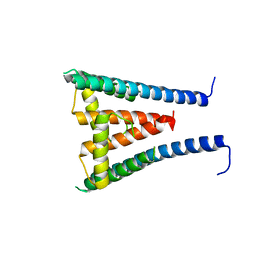 | | HUMAN SUGP1 433-586 | | Descriptor: | SURP and G-patch domain-containing protein 1 | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | DHX15 is involved in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GXL
 
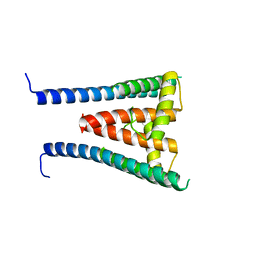 | | HUMAN SUGP1 433-577 | | Descriptor: | SURP and G-patch domain-containing protein 1 | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DHX15 is involved in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8I5X
 
 | | Structure of human Nav1.7 in complex with Vinpocetine | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5G
 
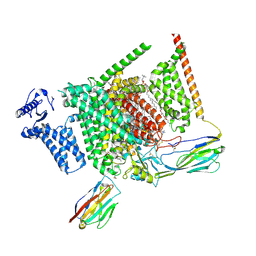 | | Structure of human Nav1.7 in complex with PF-05089771 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5B
 
 | | Structure of human Nav1.7 in complex with bupivacaine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5Y
 
 | | Structure of human Nav1.7 in complex with vixotrigine | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8J4F
 
 | | Structure of human Nav1.7 in complex with Hardwickii acid | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (4~{a}~{R},5~{S},6~{R},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{a}-trimethyl-3,4,4~{a},6,7,8-hexahydronaphthalene-1-carboxylic acid, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
5JNY
 
 | | Crystal Structure of 10E8 Fab | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, CHLORIDE ION, ... | | Authors: | Ofek, G, Kwong, P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.
Plos One, 11, 2016
|
|
5JR1
 
 | |
5NPP
 
 | | 2.22A STRUCTURE OF THIOPHENE2 AND GSK945237 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DIMETHYL SULFOXIDE, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CDM
 
 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDN
 
 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDQ
 
 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDO
 
 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDR
 
 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDP
 
 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5C0M
 
 | | Crystal structure of SGF29 tandem tudor domain in complex with a Carba containing peptide | | Descriptor: | Carba-containing peptide, GLYCEROL, SAGA-associated factor 29 homolog, ... | | Authors: | Dong, A, Xu, C, Tempel, W, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical basis for the recognition of trimethyllysine by epigenetic reader proteins.
Nat Commun, 6, 2015
|
|
5IQ7
 
 | | Crystal structure of 10E8-S74W Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8-S74W Heavy Chain, 10E8-S74W Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2869 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
5IQ9
 
 | | Crystal structure of 10E8v4 Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8v4 Heavy Chain, 10E8v4 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
4TVP
 
 | | Crystal Structure of the HIV-1 BG505 SOSIP.664 Env Trimer Ectodomain, Comprising Atomic-Level Definition of Pre-Fusion gp120 and gp41, in Complex with Human Antibodies PGT122 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Pancera, M, Zhou, T, Kwong, P.D. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and immune recognition of trimeric pre-fusion HIV-1 Env.
Nature, 514, 2014
|
|
